Search Count: 76
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2015-08-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2015-08-12 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The Parasporin-2 Bacillus Thuringiensis Toxin That Recognizes Cancer Cells
Organism: Bacillus thuringiensis serovar dakota
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2009-01-13 Classification: TOXIN Ligands: LU, GOL, EDO, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2008-03-18 Classification: HYDROLASE |
 |
Phase Transition Of Monoclinic Lysozyme Crystal Soaked In A 10% Nacl Solution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2008-03-18 Classification: HYDROLASE |
 |
Phase Transition Of Monoclinic Lysozyme Crystal Soaked In A Saturated Nacl Solution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2008-03-18 Classification: HYDROLASE |
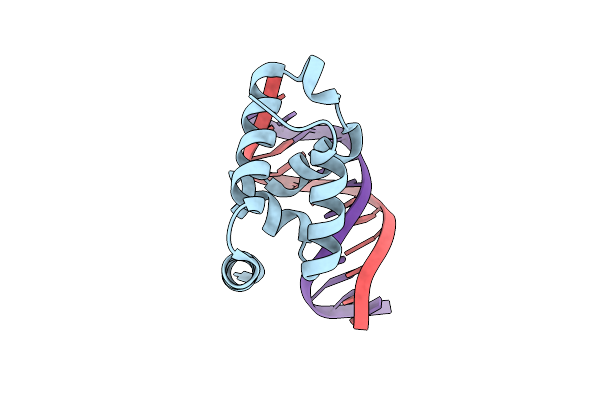 |
Crystal Structure Of The N-Terminal Cut Domain Of Satb1 Bound To Matrix Attachment Region Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-14 Classification: TRANSCRIPTION/DNA |
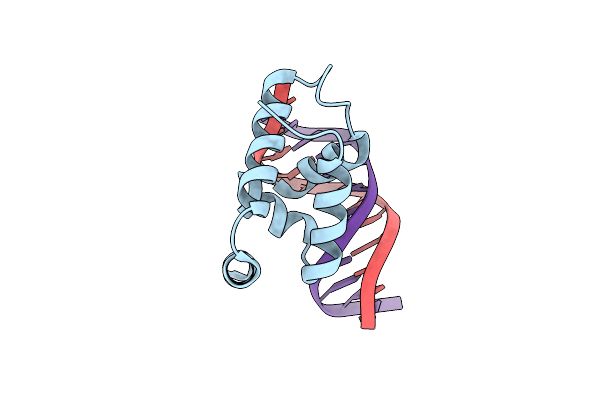 |
Crystal Structure Of The N-Terminal Cut Domain Of Satb1 Bound To Matrix Attachment Region Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-08-14 Classification: TRANSCRIPTION/DNA |
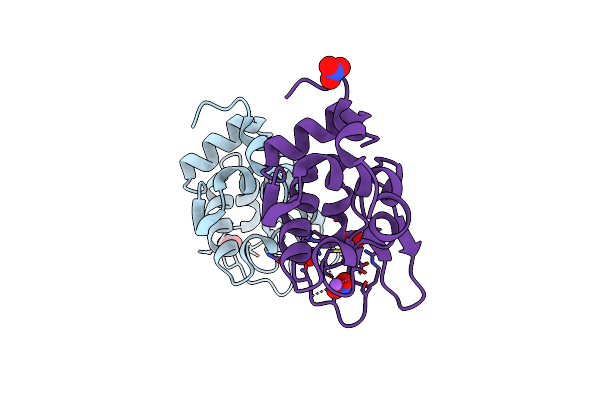 |
Monoclinic Hen Egg-White Lysozyme Crystallized At Ph4.5 Form Heavy Water Solution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: NO3, NA, DOD |
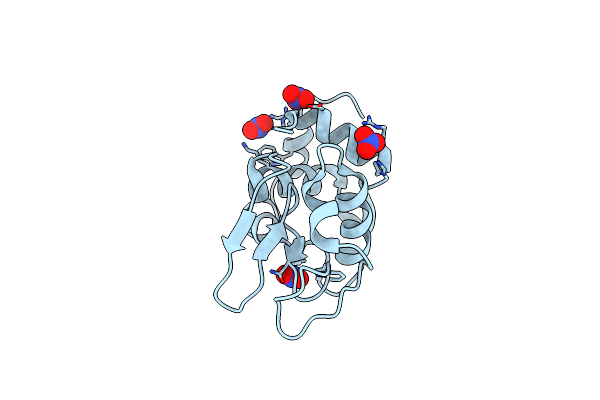 |
Transformed Monoclinic Crystal Of Hen Egg-White Lysozyme From A Heavy Water Solution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: NO3, DOD |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2006-07-11 Classification: HYDROLASE |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: SO4, IOD |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: IOD |
 |
1510-N Membrane Protease Specific For A Stomatin Homolog From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2006-05-16 Classification: HYDROLASE |
 |
Crystal Structure Of Maltohexaose-Producing Amylase From Bacillus Sp.707 Complexed With Maltopentaose.
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: GLC, CA, NA |
 |
Crystal Structure Of Maltohexaose-Producing Amylase From Bacillus Sp.707 Complexed With Maltohexaose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: GLC, CA, NA |
 |
Crystal Structure Analysis Of A Non-Toxic Crystal Protein From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2006-01-17 Classification: TOXIN |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-08-09 Classification: DNA BINDING PROTEIN |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-08-09 Classification: DNA BINDING PROTEIN |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-03-29 Classification: HYDROLASE |

