Search Count: 25
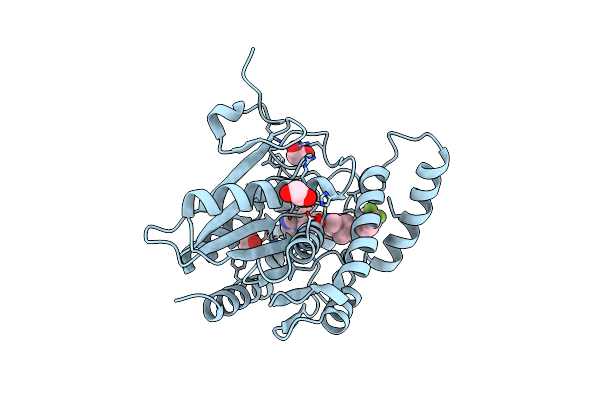 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: A1H35, EDO |
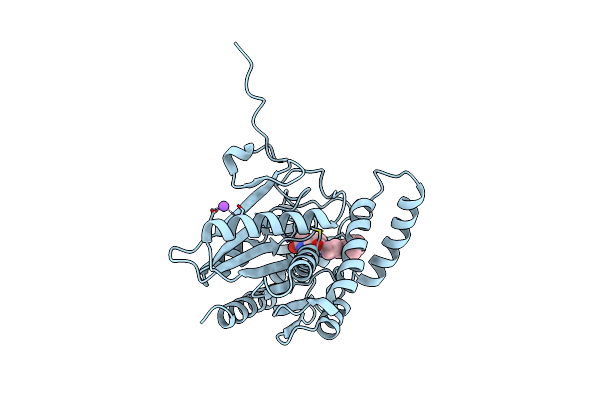 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA0, NA |
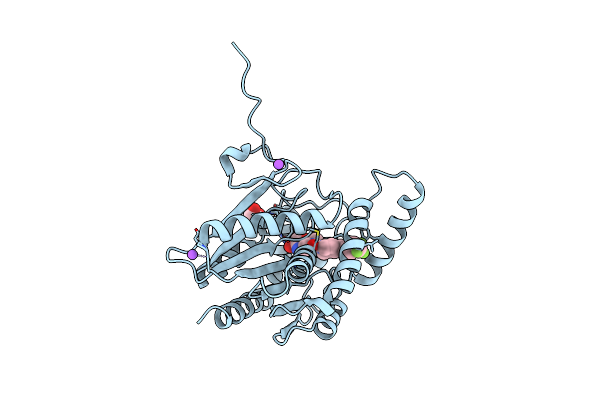 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7N
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-01-22 Classification: HYDROLASE |
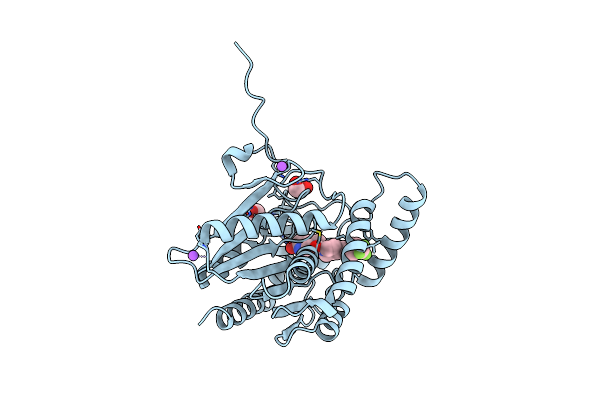 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7M
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA2, NA, EDO |
 |
Crystal Structure Of Human Monoacylglycerol Lipase In Complex With Compound 7I
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: A1IA1, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: A1IG3, EDO |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-12-27 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG, ZN, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
 |
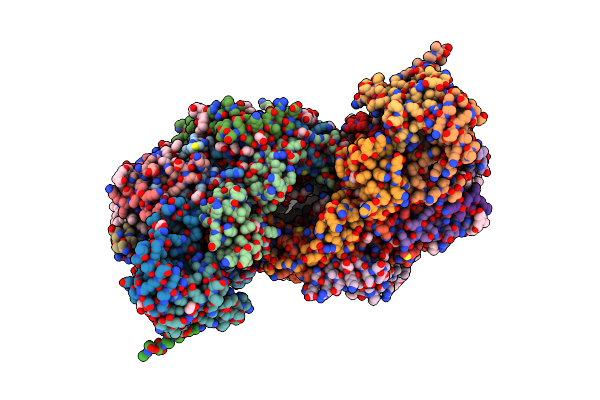
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate Of E At 1.76 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, PSC, PER, EDO, TGL, CUA, CHD, DMU, CDL, PEK, ZN, PO4 |
 |
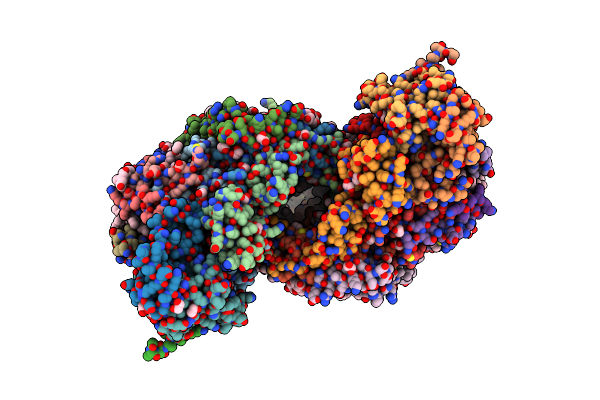
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate Of O At 1.84 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, TGL, PGV, PER, PSC, EDO, DMU, CUA, CHD, PEK, CDL, ZN, PO4 |
 |
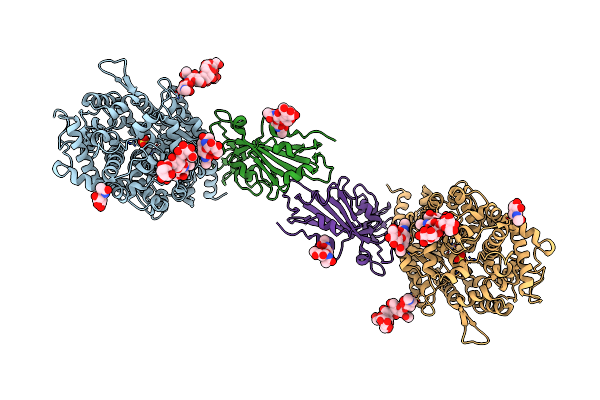
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate, Io10, At 1.74 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, OXY, EDO, DMU, TGL, CUA, PSC, CHD, PEK, CDL, ZN, PO4 |
 |
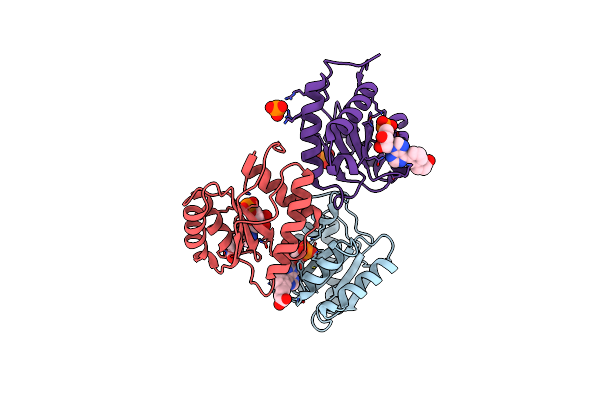
Structure Of Sars-Cov-2 Spike Receptor-Binding Domain Complexed With High Affinity Ace2 Mutant 3N39
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-12-23 Classification: VIRAL PROTEIN Ligands: NAG, ZN, SO4 |
 |
Crystal Structure Of Ago2 Mid Domain In Complex With 6-(3-(2-Carboxyethyl)Phenyl)Purine Riboside Monophosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-25 Classification: RNA BINDING PROTEIN Ligands: K2R, PO4 |
 |
Crystal Structure Of Ago2 Mid Domain In Complex With 8-Br-Adenosin-5'-Monophosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-25 Classification: RNA BINDING PROTEIN Ligands: 8BR |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans In Complex With Fsa, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, D1D |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With 7 Additional Mutations, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-02-01 Classification: ELECTRON TRANSPORT Ligands: FES |

