Search Count: 706
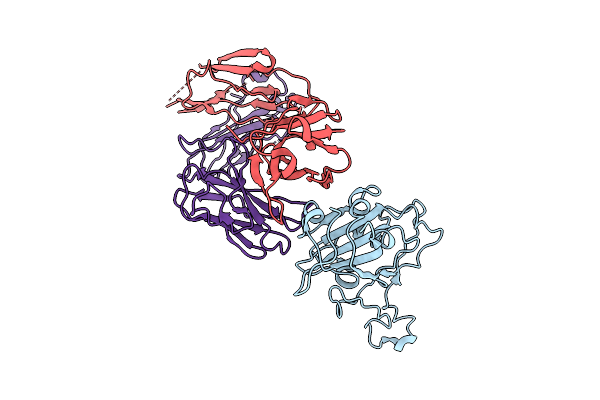 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
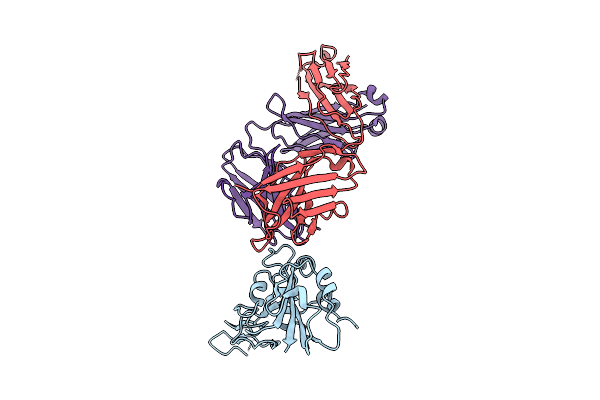
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
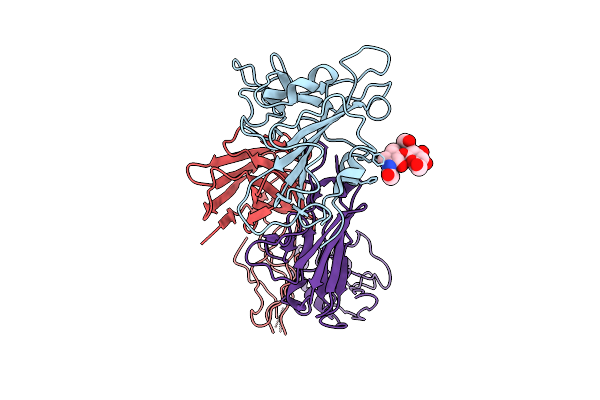
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
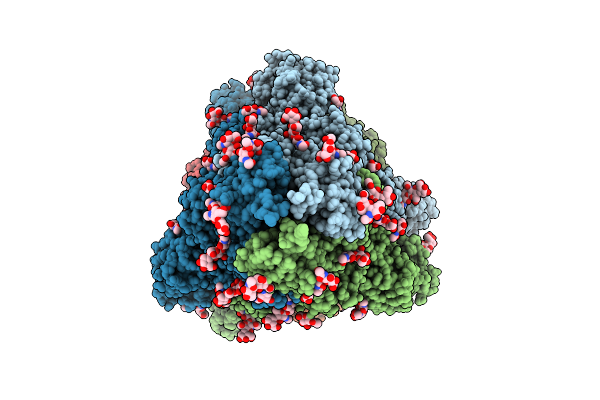
 |
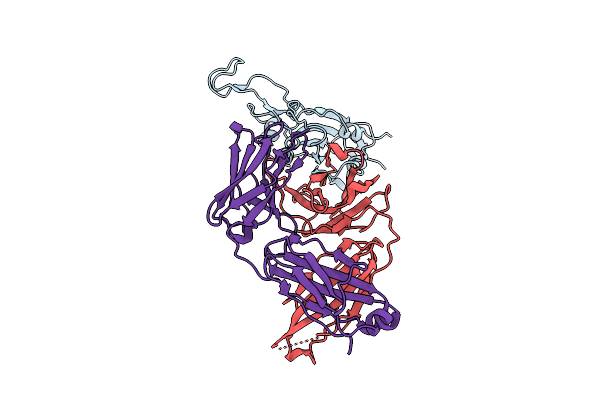
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
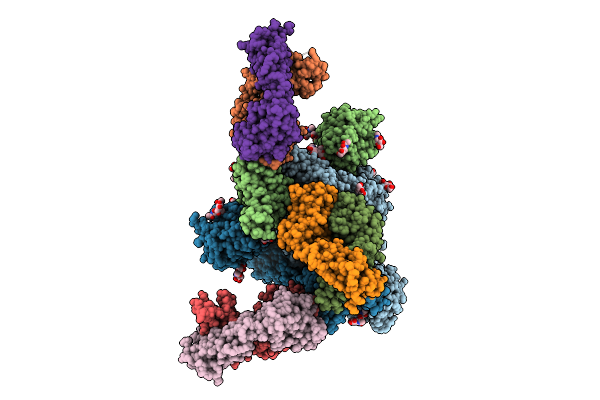
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
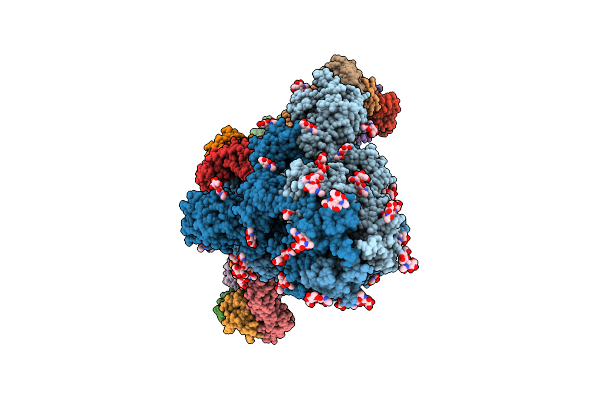
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
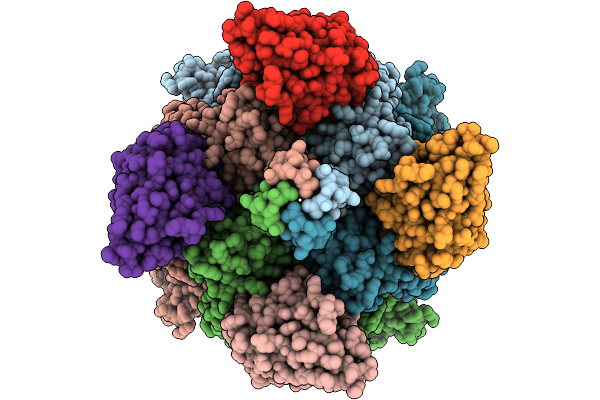
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Camelus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, A1L5P |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, A1L5Q |
 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: CA, ZN, A1L5I |
 |
The Glycoprotein E Of Varicella-Zoster Virus In Complex With Wll-1/Wll-28 Fab
Organism: Varicella-zoster virus (strain oka vaccine), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Fusobacterium Nucleatum Cbpf In Complex With Human Ceacam5
Organism: Fusobacterium nucleatum, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL ADHESION |
 |
Cryo-Em Structure Of Fusobacterium Nucleatum Cbpf In Complex With Human Ceacam1
Organism: Fusobacterium nucleatum, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL ADHESION |
 |
Organism: Epilithonimonas lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE Ligands: C2E, CA |
 |
Organism: Epilithonimonas lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE Ligands: C2E |
 |
Organism: Bacillus phage spo1
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRUS |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |

