Search Count: 344
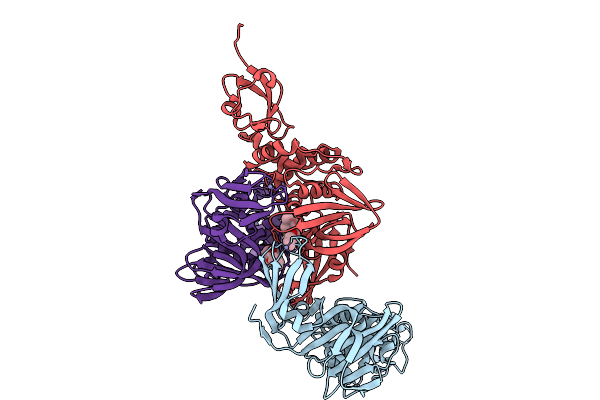 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1CLE, ZN |
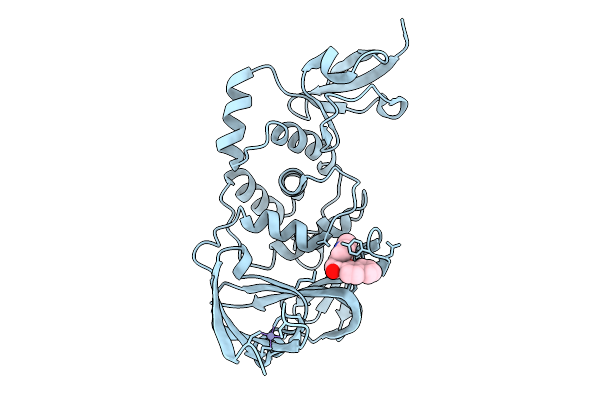 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLF |
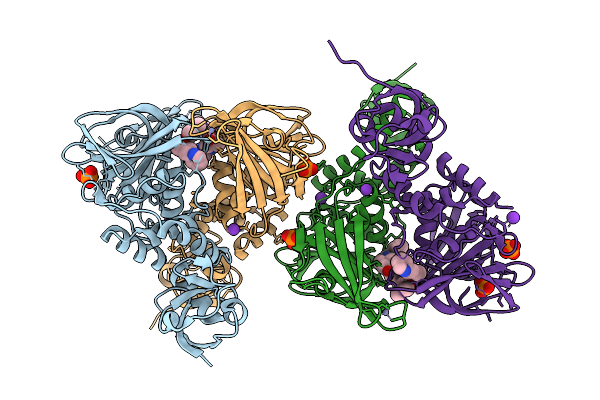 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLD, PO4, K |
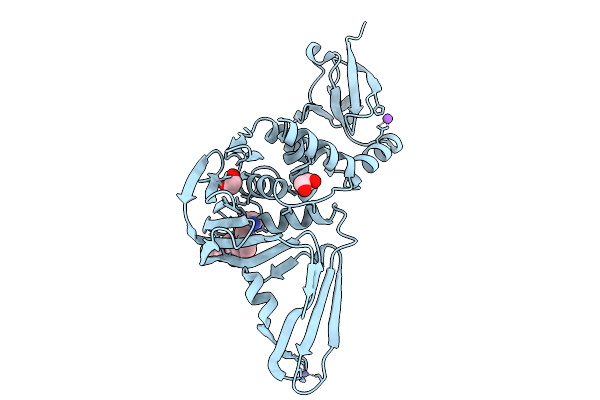 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, EDO, A1CLN, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLP, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLO, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, EDO, A1CLL |
 |
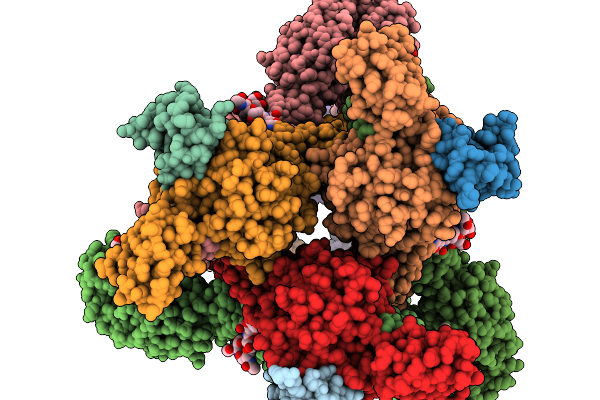
Organism: Homo sapiens, Mus musculus, Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS Ligands: CA |
 |
Organism: Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
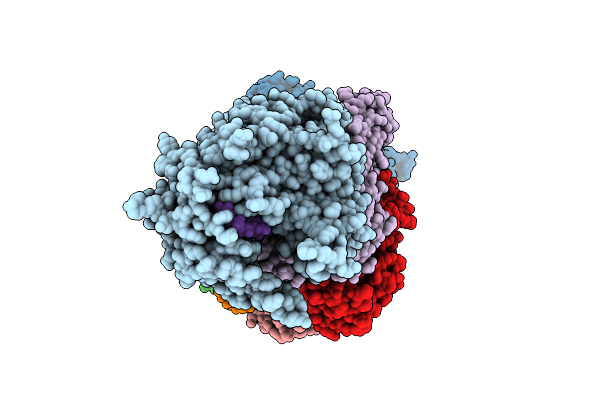 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: MN |
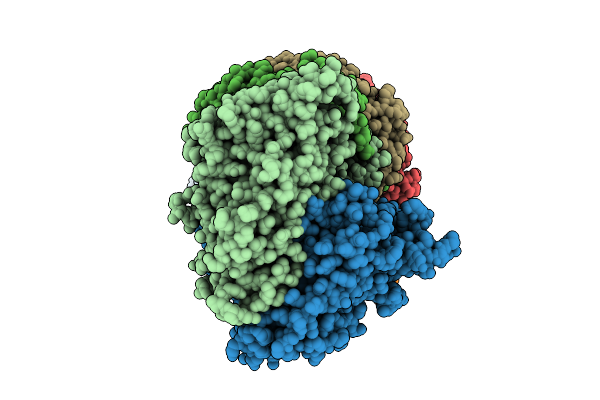 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
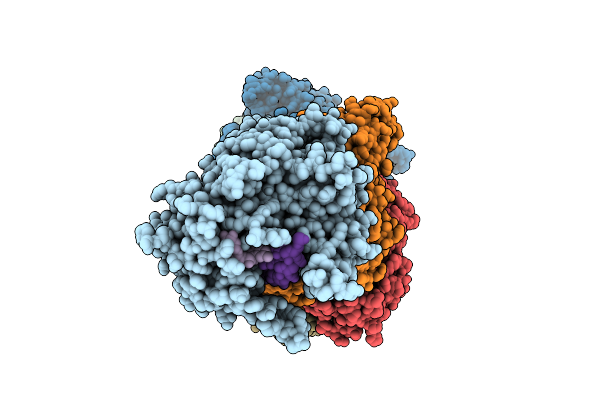 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL3, TRS, MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL0, MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL0 |
 |
Organism: Clostridium botulinum (strain hall / atcc 3502 / nctc 13319 / type a)
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: MLT |
 |
Organism: Clostridium botulinum (strain hall / atcc 3502 / nctc 13319 / type a)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: SAH |
 |
Organism: Clostridium botulinum (strain hall / atcc 3502 / nctc 13319 / type a)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: A1EL0, GOL |

