Search Count: 108
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With One Ace2 Receptor Binding (Rb1) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With Two Ace2 Receptors Binding (Rb2) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With Three Ace2 Receptors Binding (Rb3) In Prefusion Conformation (Focused Refinement Of Ntd-Sd1-Rbd-Ace2)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
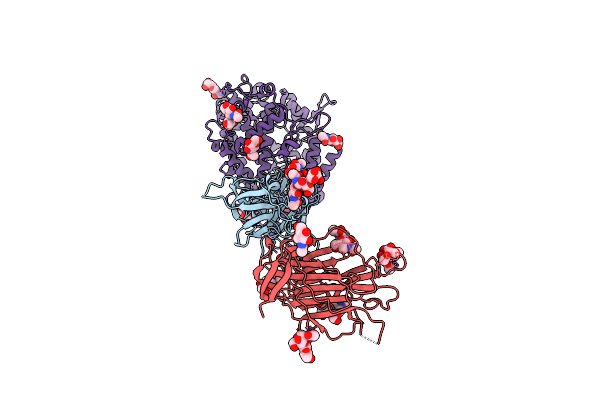
Cryo-Em Structure Of Sars-Cov-2 D614G S With Three Ace2 Receptors Binding (Rb3) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
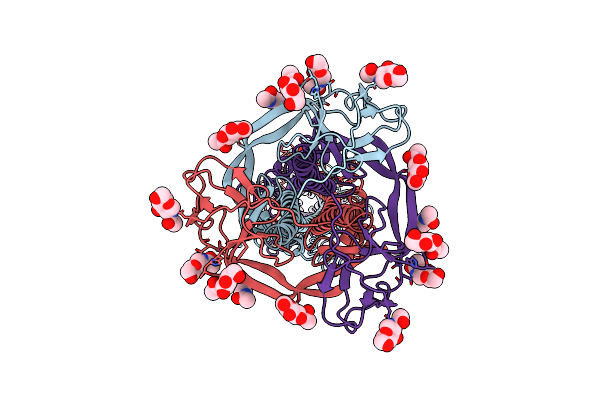
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of Ntd-Sd1-Rbd-Ace2)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
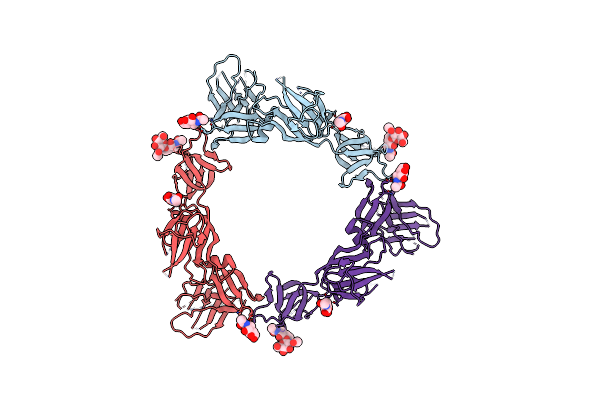
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of Intact S2)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
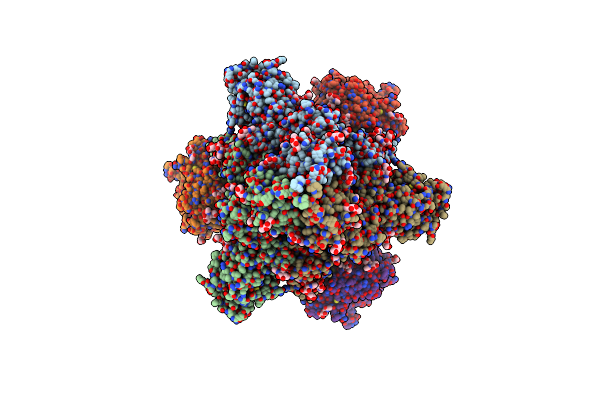
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of S-Bottom)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HTDROLASE Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: A1ADE, SO4, GDP |
 |
Organism: Aquifex aeolicus vf5, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: TRANSFERASE Ligands: A1AI1 |
 |
Organism: Lama glama, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: TRANSFERASE Ligands: A1AI2 |
 |
Structure Of Xbb Spike Protein (S) Dimer-Trimer In Complex With Bispecific Antibody G7-Fc At 3.75 Angstroms Resolution.
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSPORT PROTEIN Ligands: MG, ADP |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSPORT PROTEIN Ligands: MG, ADP |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSPORT PROTEIN Ligands: MG, ADP |

