Search Count: 38
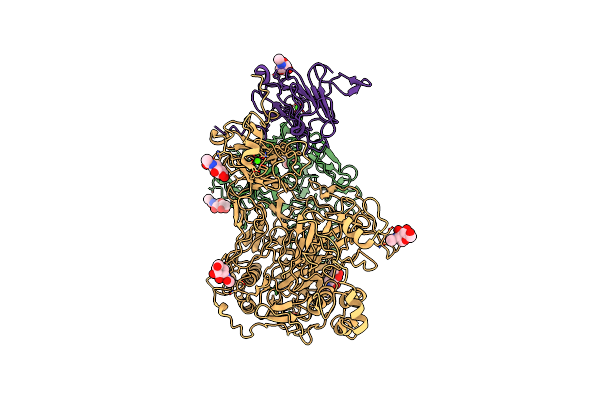 |
Structure Of The Mouse Fcgbp Dimer Protein In Its Semiextended Conformation
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2025-01-08 Classification: STRUCTURAL PROTEIN Ligands: NAG, CA |
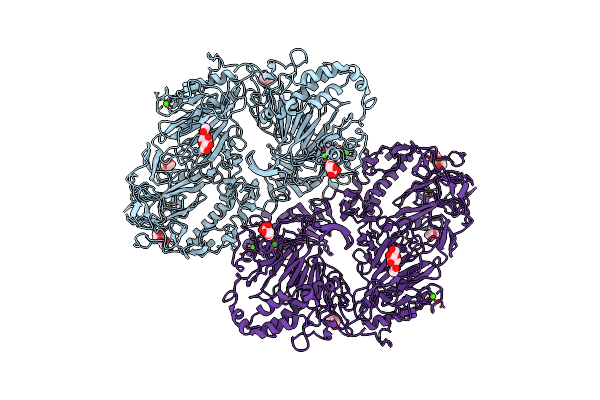 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-01-08 Classification: STRUCTURAL PROTEIN Ligands: NAG, CA |
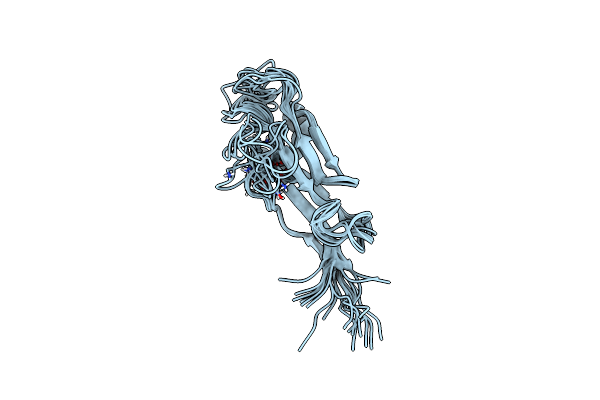 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-04-17 Classification: STRUCTURAL PROTEIN Ligands: CA |
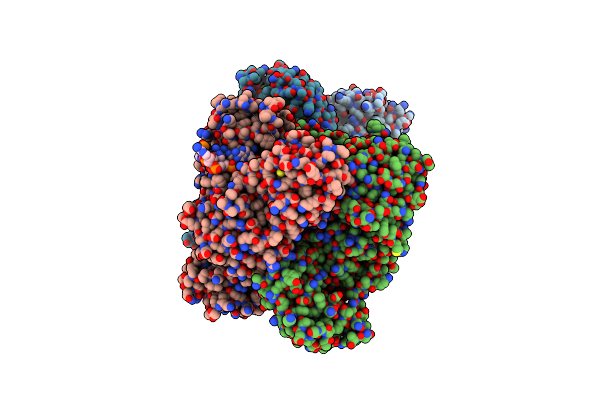 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-04-03 Classification: ANTIVIRAL PROTEIN Ligands: MG |
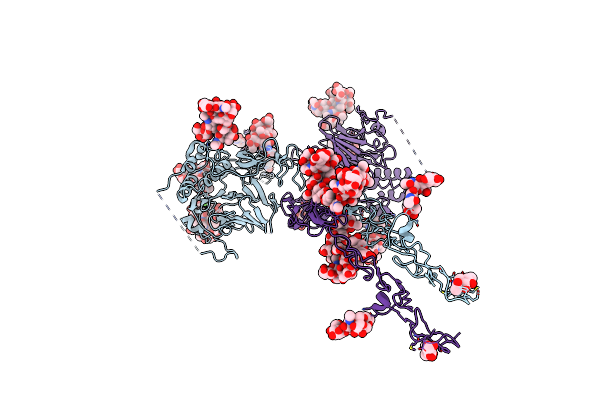 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: STRUCTURAL PROTEIN Ligands: NAG, CA |
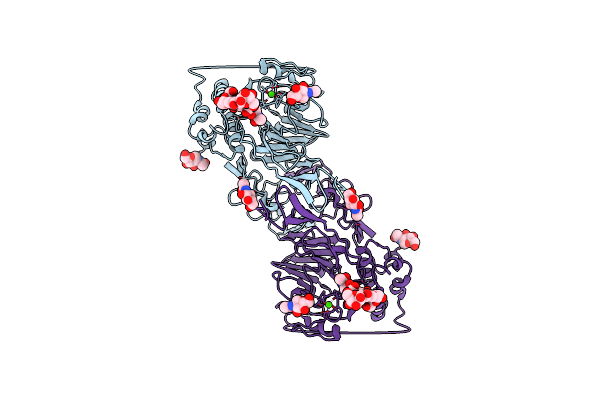 |
Structure Of The Mucin-2 Cterminal Domains: Vwcn To Til Domains With A C2 Symmetry
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: STRUCTURAL PROTEIN Ligands: CA, NAG |
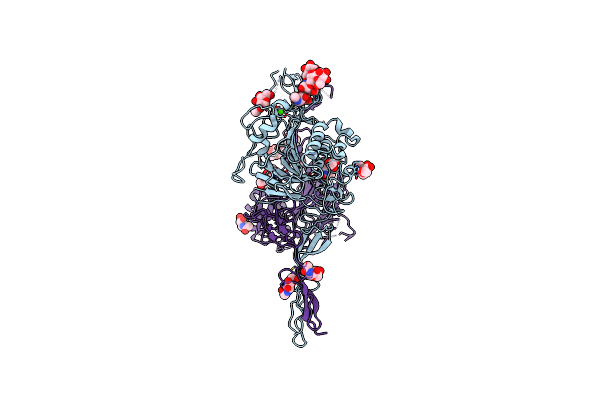 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: STRUCTURAL PROTEIN Ligands: CA, NAG |
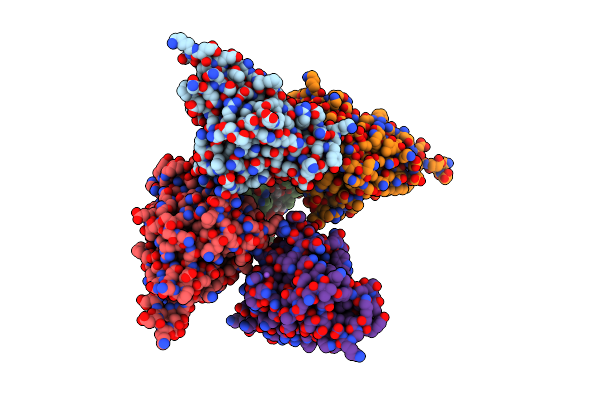 |
Organism: Streptococcus suis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-08-19 Classification: SUGAR BINDING PROTEIN Ligands: GOL, NA |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: RIBOSOMAL PROTEIN Ligands: ATP |
 |
Single-Particle Cryo-Em Of Co-Translational Folded Adr1 Domain Inside The E. Coli Ribosome Exit Tunnel.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:4.80 Å Release Date: 2015-09-16 Classification: TRANSLATION Ligands: ZN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2013-03-20 Classification: RNA/antibiotic Ligands: AM2 |
 |
Modelling Of The Complex Between Subunits Bchi And Bchd Of Magnesium Chelatase Based On Single-Particle Cryo-Em Reconstruction At 7.5 Ang
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:7.50 Å Release Date: 2010-11-10 Classification: LIGASE |
 |
Crystal Structure Of B. Subtilis Ferrochelatase With Cobalt Bound At The Active Site
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-11-10 Classification: LYASE Ligands: CO, MG, CL |
 |
Crystal Structure Of The Tyr13Met Variant Of Bacillus Subtilis Ferrochelatase
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-31 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of H183C Bacillus Subtilis Ferrochelatase In Complex With Deuteroporphyrin Ix 2,4-Disulfonic Acid Dihydrochloride
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-06-17 Classification: LYASE Ligands: MG, H01 |
 |
Crystal Structure Of Bacillus Subtilis Ferrochelatase In Complex With Deuteroporphyrin Ix 2,4-Disulfonic Acid Dihydrochloride
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-22 Classification: TRANSFERASE Ligands: MG, H01 |
 |
Crystal Structure Of The His183Ala Variant Of Bacillus Subtilis Ferrochelatase In Complex With N-Methyl Mesoporphyrin
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2008-04-22 Classification: LYASE Ligands: MG, H02 |
 |
Nmr Solution Structure Of A Minimal Transmembrane Beta-Barrel Platform Protein
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Lys87Ala Mutant Variant Of Bacillus Subtilis Ferrochelatase
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2007-01-16 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of The His183Ala Mutant Variant Of Bacillus Subtilis Ferrochelatase
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-01-16 Classification: LYASE Ligands: FE2, MG |

