Search Count: 14
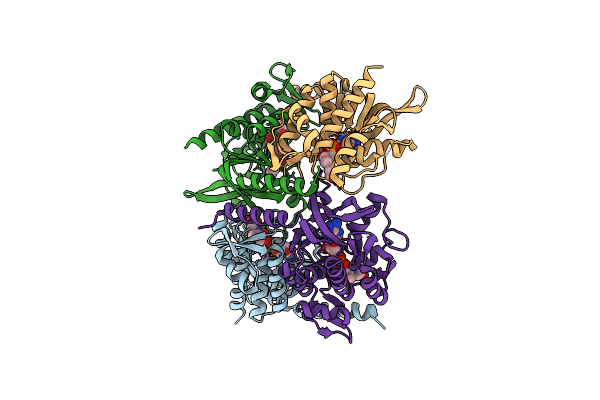 |
Organism: Thermovibrio ammonificans
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-09-01 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, 48H |
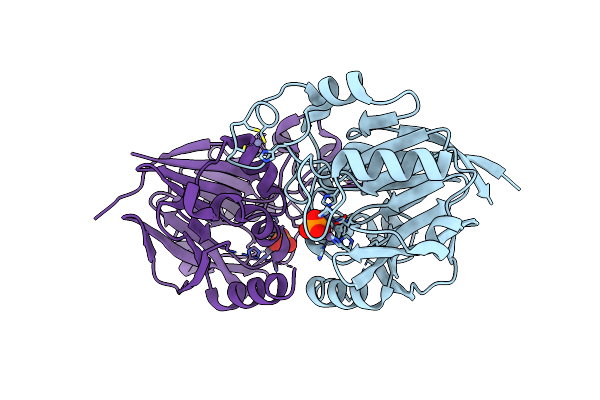 |
Organism: Koribacter versatilis (strain ellin345)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: MN, PO4, ZN |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase From Bacillus Thuringiensis Involved In Dispatching The Ubiquitous Radical Sam Enzyme Byproduct 5-Deoxyribose
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-07-04 Classification: LYASE Ligands: SO4, MN |
 |
Crystal Structure Of Deoxyribose-Phosphate Aldolase Bound With Dhap From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-07-04 Classification: LYASE Ligands: MN, 13P |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2016-03-30 Classification: TRANSFERASE Ligands: MN, PO4, CL, EDO |
 |
Crystal Structure Of The Homocysteine Methyltransferase Mmum From Escherichia Coli, Oxidized Form
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: CL |
 |
Crystal Structure Of The Homocysteine Methyltransferase Mmum From Escherichia Coli, Metallated Form
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: ZN, BME, HCS |
 |
Crystal Structure Of The Homocysteine Methyltransferase Mmum From Escherichia Coli, Apo Form
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of H5 Hemagglutinin Mutant (N158D, N224K And Q226L) From The Influenza Virus A/Viet Nam/1203/2004 (H5N1)
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2013-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
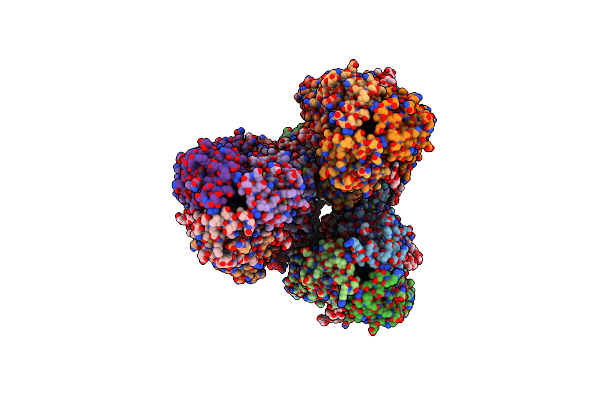 |
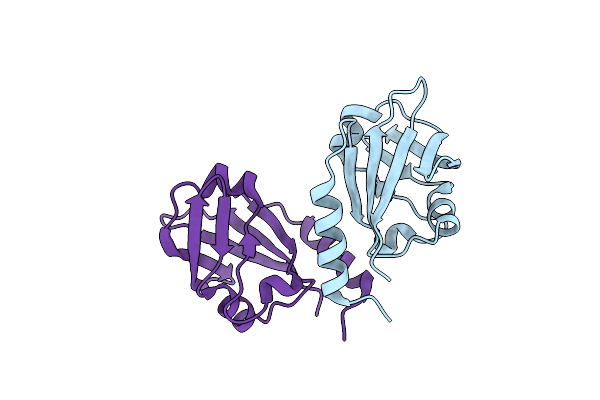
Crystal Structure Of Aerosol Transmissible Influenza H5 Hemagglutinin Mutant (N158D, N224K, Q226L And T318I) From The Influenza Virus A/Viet Nam/1203/2004 (H5N1)
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-11-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-12-19 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Organism: Momordica cochinchinensis
Method: SOLUTION NMR Release Date: 2012-08-08 Classification: HYDROLASE INHIBITOR |
 |
 |
Atomic Model Of Bovine Tric Cct2(Beta) Subunit Derived From A 4.0 Angstrom Cryo-Em Map
|

