Search Count: 156
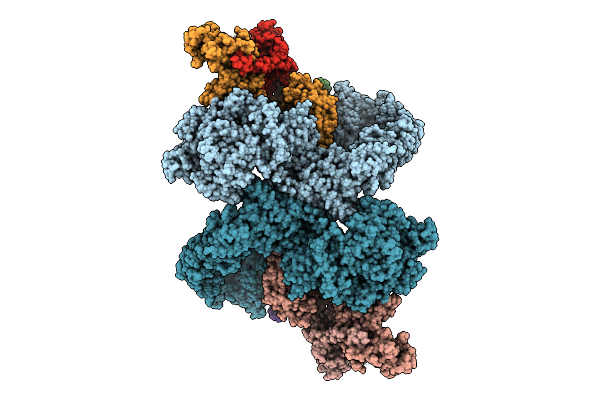 |
Cryo-Em Structure Of The Pi4Ka Complex Bound To An Efr3 Interfering Nanobody (F3In)
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.54 Å Release Date: 2025-12-03 Classification: SIGNALING PROTEIN |
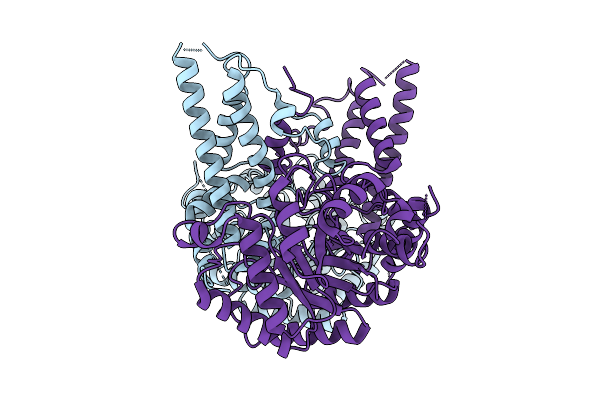 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE |
 |
Organism: Trichoplax adhaerens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, ACT, GOL |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, SO4, EDO |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With A Nanobody
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: GOL, NAG |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN |
 |
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: ACT, IMD, GOL, SO4, NAG |
 |
Crystal Structure Of Lotus Japonicus Chip13 Extracellular Domain In Complex With Chitooctaose
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: IMD, EDO |
 |
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: NAG, EDO, MAN |
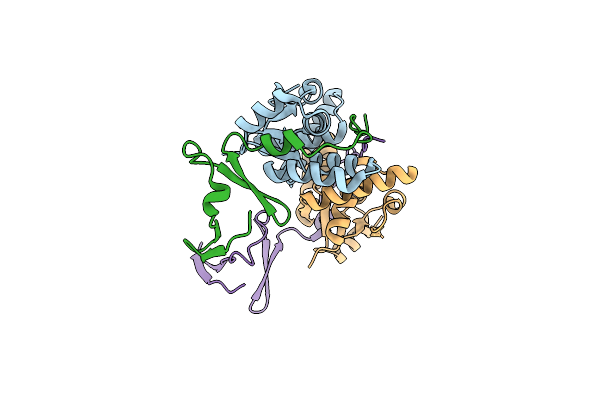 |
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-12-18 Classification: TOXIN |
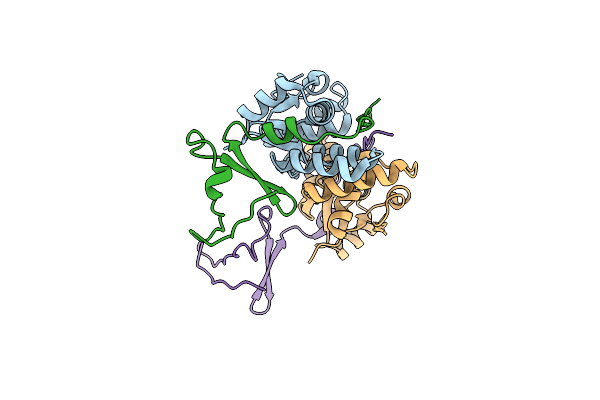 |
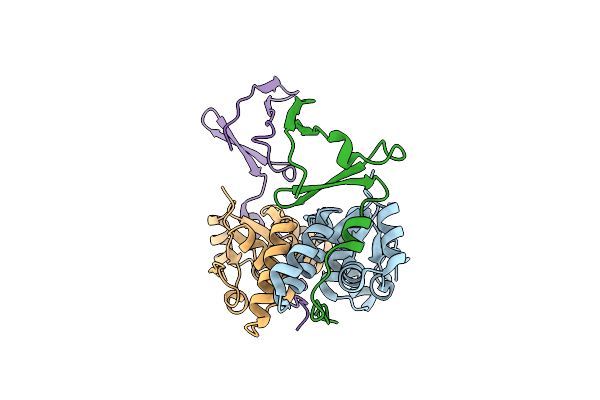
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N, A13P, L16R)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
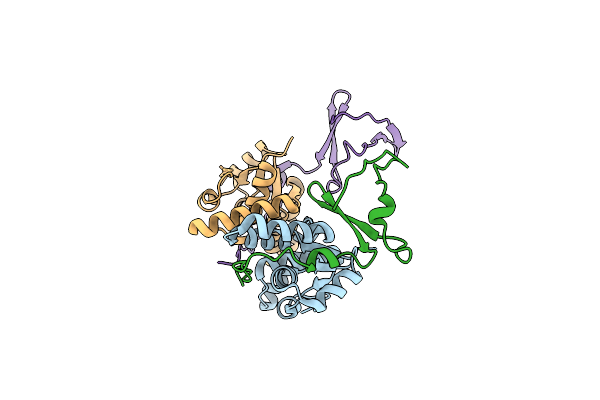
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
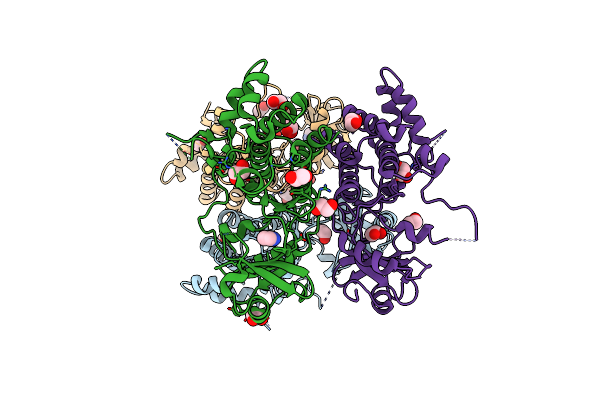
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb V5E)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
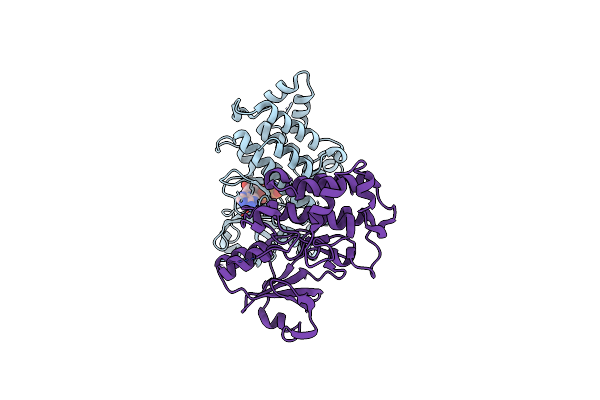
Organism: Lotus japonicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-11-27 Classification: PLANT PROTEIN Ligands: EDO, IMD |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-27 Classification: PLANT PROTEIN Ligands: ANP |
 |
Organism: Lotus japonicus, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-11-06 Classification: PLANT PROTEIN Ligands: PEG, BME, ACT |

