Search Count: 142
 |
Structural Basis Of Pseudomonas Fapc Biofilm-Forming Functional Amyloid Formation
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: STRUCTURAL PROTEIN |
 |
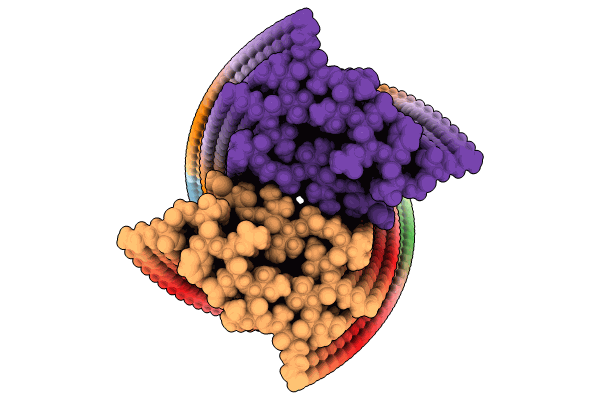
Crystal Structure Of Glun1/Glun2A Agonist-Binding Domains In Complex With 7Cka And Glutamate
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-03-12 Classification: SIGNALING PROTEIN Ligands: CKA, GLU |
 |
Structure Of Biofilm-Forming Functional Amyloid Psma1 From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: STRUCTURAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: SPLICING |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: CU, NAG, AKR, 2HA, CL |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: RNA |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-31 Classification: OXIDOREDUCTASE Ligands: CU, CL, SO4, ACT |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: ACT, CU, CL, SO4 |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
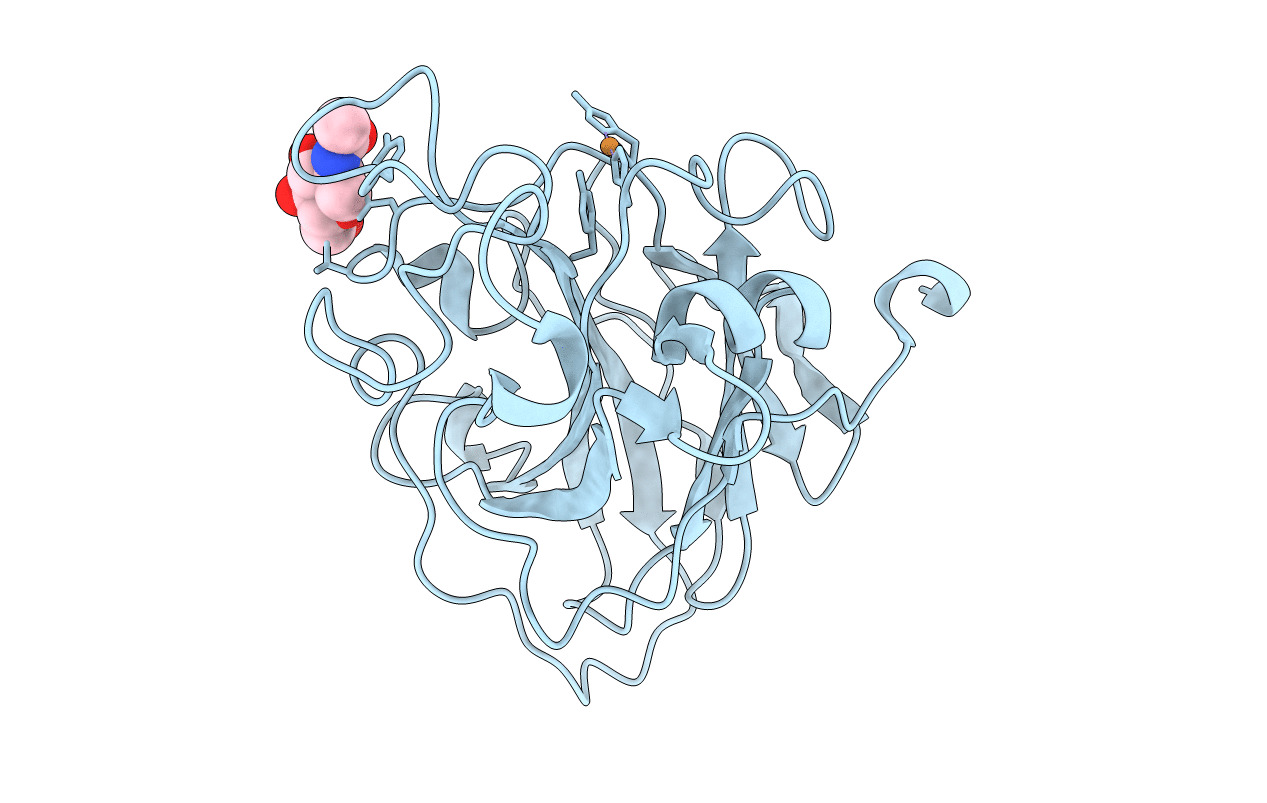 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
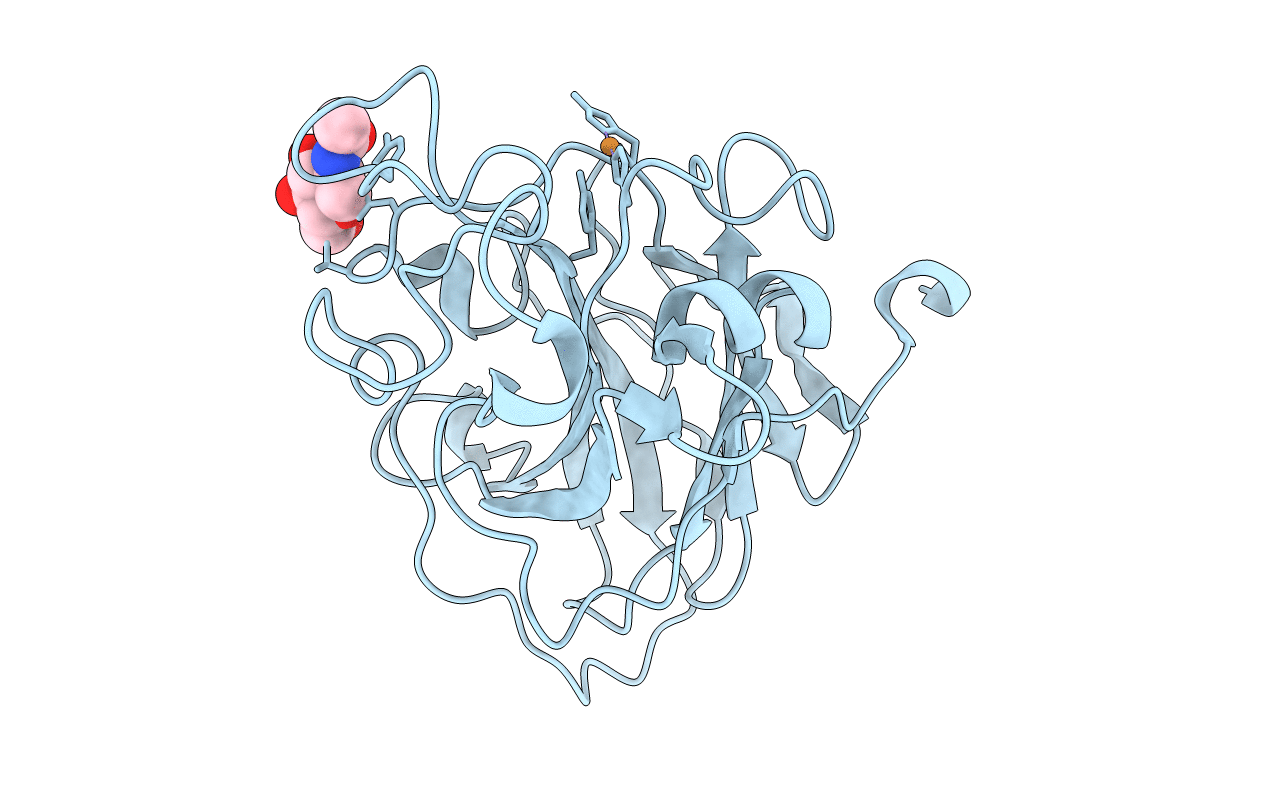 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
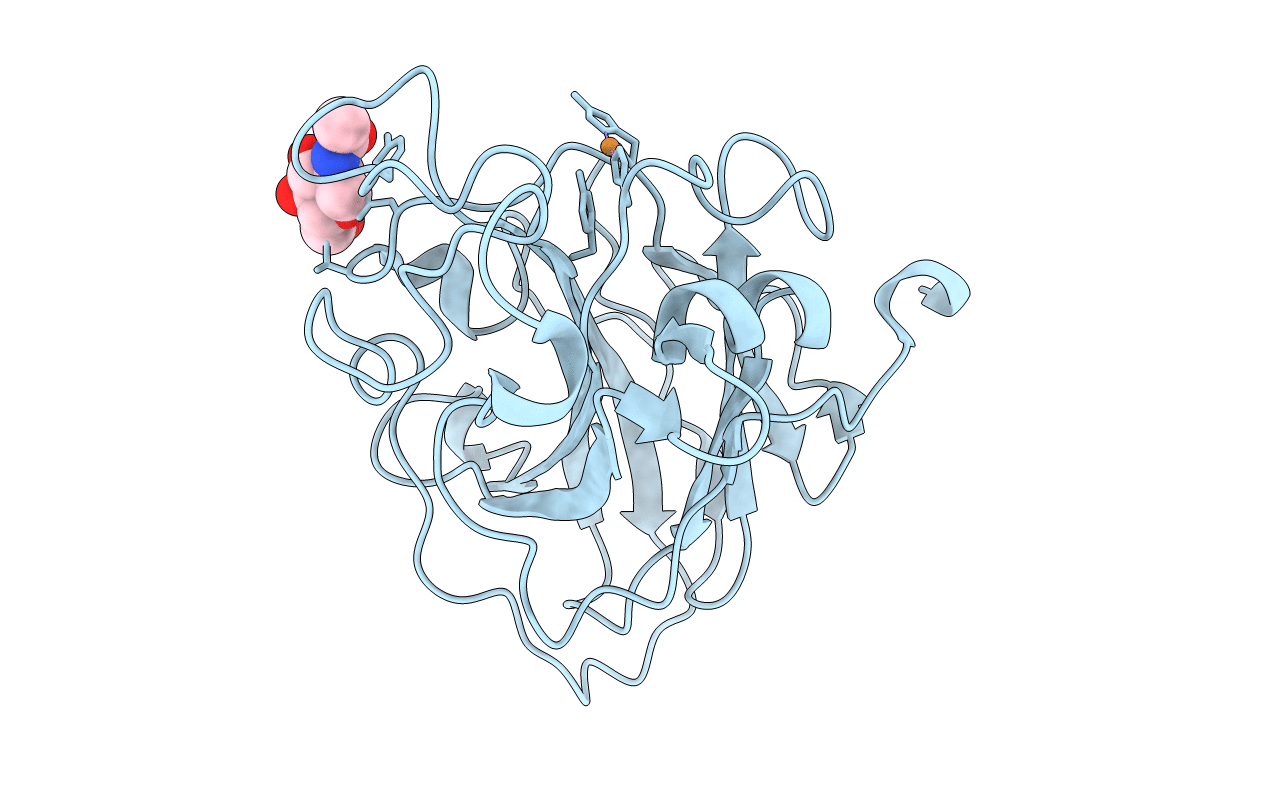 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
 |
Structure Of An Lpmo, Collected From Serial Synchrotron Crystallography Data.
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: SO4, CU, CL |

