Search Count: 13
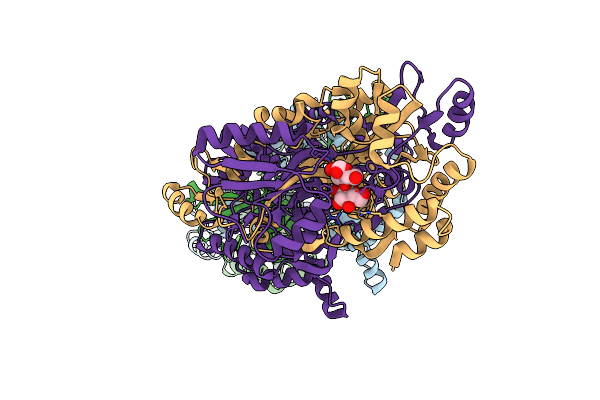 |
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2021-08-04 Classification: HORMONE |
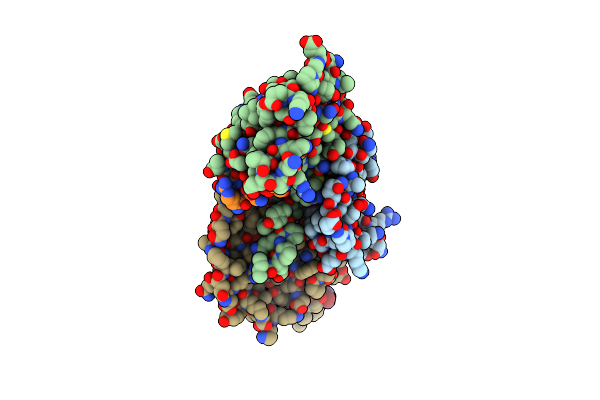 |
Crystal Structure Of The Human Calcitonin Receptor Ectodomain In Complex With A Truncated Salmon Calcitonin Analogue
Organism: Homo sapiens, Oncorhynchus gorbuscha
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-05-11 Classification: HORMONE |
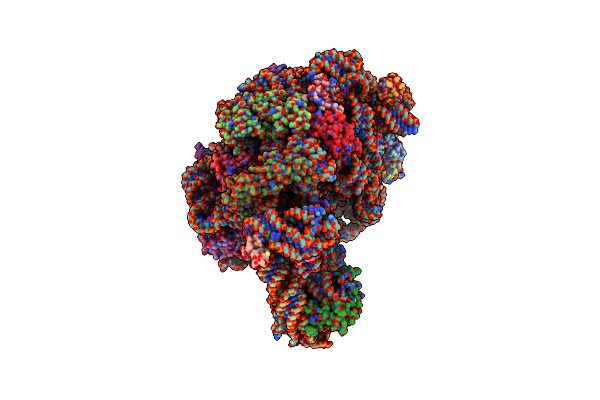 |
Crystal Structure Of Ccdap-Puromycin Bound At The Peptidyl Transferase Center Of The 50S Ribosomal Subunit
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2003-10-07 Classification: RIBOSOME Ligands: MG, K, NA, CL, PO4, PUY, CD |
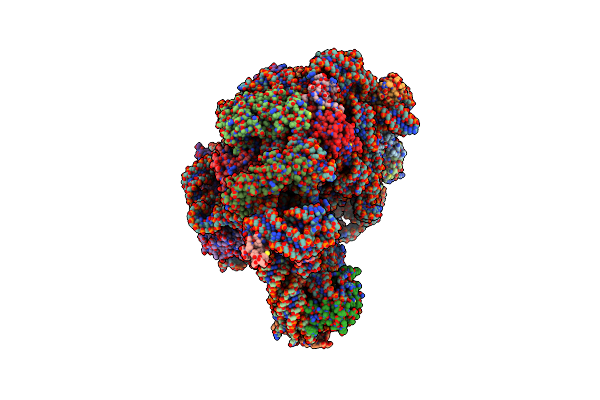 |
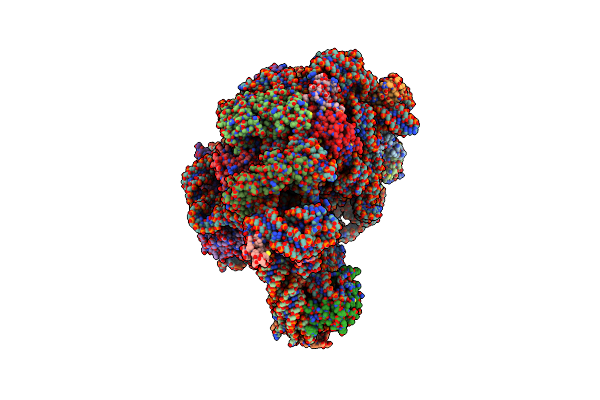
Crystal Structure Of Minihelix With 3' Puromycin Bound To A-Site Of The 50S Ribosomal Subunit.
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2003-10-07 Classification: RIBOSOME Ligands: MG, K, NA, CL, PPU, CD |
 |
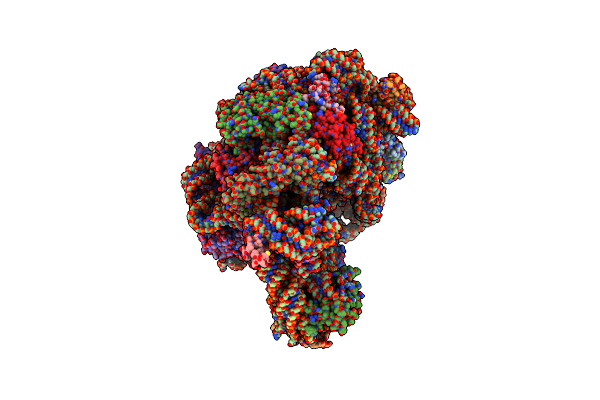
Crystal Structure Of Cc-Puromycin Bound To The A-Site Of The 50S Ribosomal Subunit
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2003-10-07 Classification: RIBOSOME Ligands: MG, K, NA, CL, PPU, CD |
 |
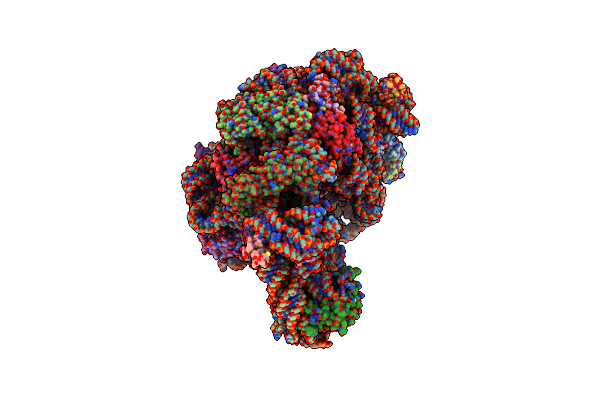
Crystal Structure Of Cca-Phe-Cap-Biotin Bound Simultaneously At Half Occupancy To Both The A-Site And P-Site Of The The 50S Ribosomal Subunit.
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2003-10-07 Classification: RIBOSOME Ligands: MG, K, NA, CL, PHA, CD |
 |
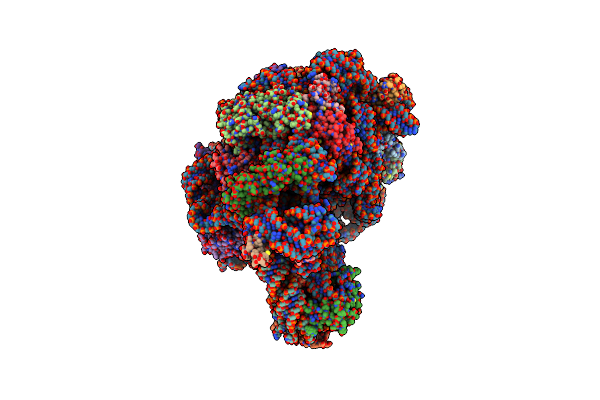
Co-Crystal Structure Of Cca-Phe-Caproic Acid-Biotin And Sparsomycin Bound To The 50S Ribosomal Subunit
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-09-06 Classification: RIBOSOME Ligands: MG, NA, K, CL, SPS, PHA, ACA, CD |
 |
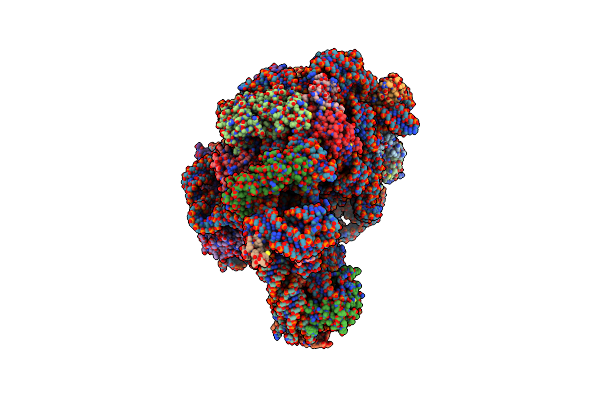
Co-Crystal Structure Of Carbomycin A Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-07-19 Classification: RIBOSOME Ligands: CAI, MG, NA, CL, K, CD |
 |
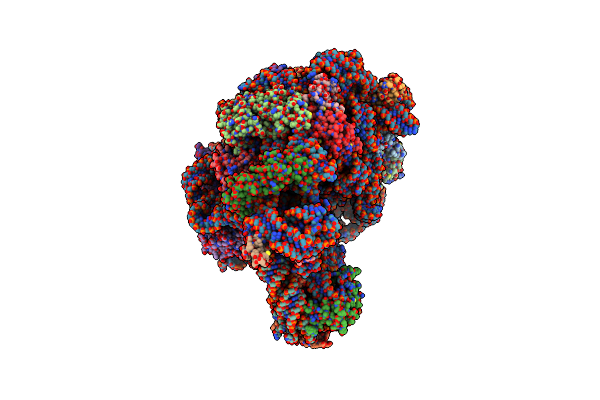
Co-Crystal Structure Of Tylosin Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-07-19 Classification: RIBOSOME Ligands: TYK, MG, NA, CL, K, CD |
 |
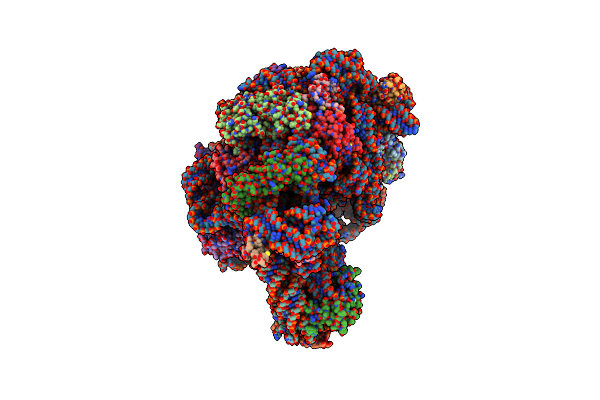
Co-Crystal Structure Of Spiramycin Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-07-19 Classification: RIBOSOME Ligands: SPR, MG, NA, CL, K, CD |
 |
Co-Crystal Structure Of Azithromycin Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-07-19 Classification: RIBOSOME Ligands: ZIT, MG, K, NA, CL, CD |
 |
The Haloarcula Marismortui 50S Complexed With A Pretranslocational Intermediate In Protein Synthesis
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2002-02-22 Classification: RIBOSOME Ligands: MG, K, NA, CL, PPU, PHA, ACA, BTN, CD |
 |
Organism: Human poliovirus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-09-16 Classification: NUCLEOTIDYLTRANSFERASE Ligands: CA |

