Search Count: 15
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
The Condensation Domain Of Surfactin A Synthetase C Variant 18B In Space Group P212121
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
The Condensation Domain Of Surfactin A Synthetase C Variant 18B In Space Group P43212
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
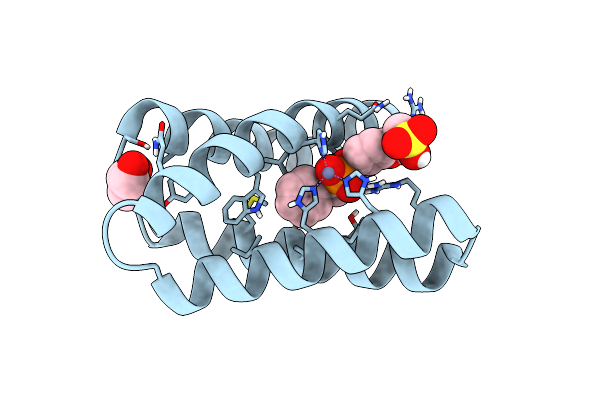 |
Structure Of The Engineered Metalloesterase Mid1Sc10 Complexed With A Phosphonate Transition State Analogue
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2018-12-12 Classification: De Novo Protein/Hydrolase Ligands: ZN, 9RQ, GOL |
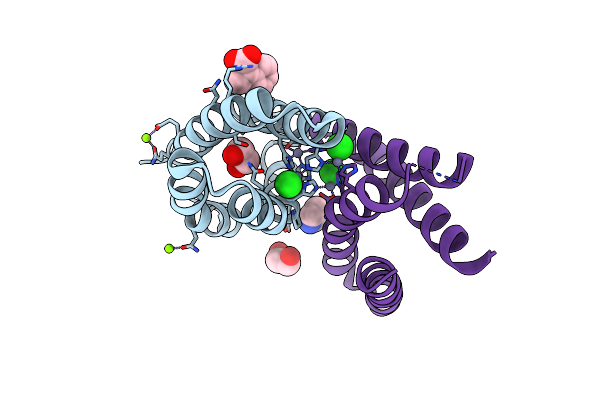 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2018-12-12 Classification: DE NOVO PROTEIN HYDROLASE Ligands: ZN, CL, MG, EDO, IMD, 9RW |
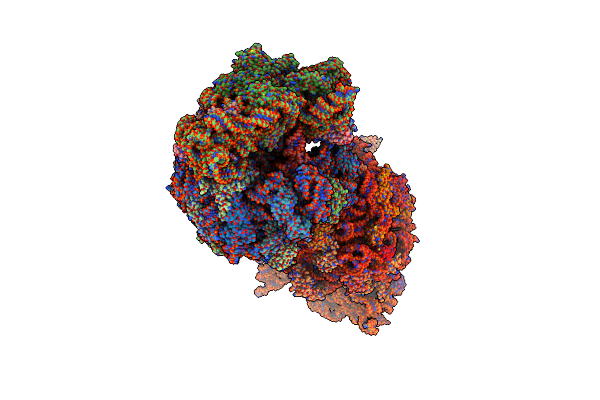 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Methymycin And Bound To Mrna And A-, P- And E-Site Trnas At 2.7A Resolution
Organism: Escherichia coli, Enterobacteria phage t4, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-14 Classification: RIBOSOME Ligands: MG, MT9, K, ZN, SF4 |
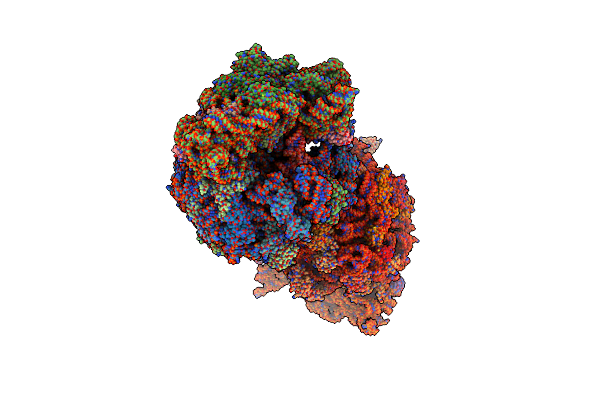 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Pikromycin And Bound To Mrna And A-, P- And E-Site Trnas At 2.6A Resolution
Organism: Escherichia coli, Enterobacteria phage t4, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-02-14 Classification: RIBOSOME Ligands: MG, K, AQJ, ZN, SF4 |
 |
Crystal Structure Of An Engineered Tyca Variant In Complex With An O-Propargyl-Beta-Tyr-Amp Analog
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-12-13 Classification: LIGASE Ligands: 8Q2, SO4, BTB |
 |
Crystal Structure Of An Engineered Tyca Variant In Complex With An Beta-Phe-Amp Analog
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-12-13 Classification: LIGASE Ligands: SO4, BTB, 8PZ |
 |
Pikc D50N Mutant Bound To The 10-Dml Analog With The 3-(N,N-Dimethylamino)Ethanoate Anchoring Group
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2014-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, Z18 |
 |
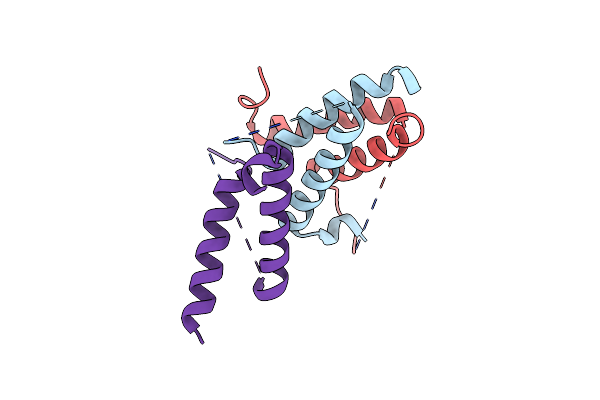
Structure Of A Class 2 Docking Domain Complex From Modules Curg And Curh Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-01-29 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Structure Of A Class 2 Docking Domain Complex From Modules Curk And Curl Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-01-29 Classification: PROTEIN BINDING |
 |
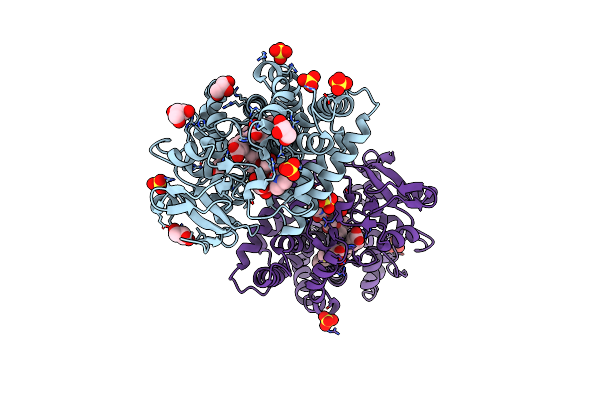
Structure Of A Ketosynthase-Acyltransferase Di-Domain From Module Curl Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-01-29 Classification: TRANSFERASE Ligands: CA |
 |
Pikc D50N Mutant Bound To The 10-Dml Analog With The 3-(N,N- Dimethylamino)Propanoate Anchoring Group
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, QLE, GOL |

