Search Count: 20
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-10 Classification: PROTEIN BINDING Ligands: B3P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-01-10 Classification: PROTEIN BINDING |
 |
Crystal Structure Of 14-3-3 In Complex With Pyrinps242 And A Protein/Peptide Interface Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-01-10 Classification: PROTEIN BINDING Ligands: 463, PE5, MG, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-07-26 Classification: PROTEIN BINDING Ligands: MG |
 |
N-Terminal Bromodomain Of Human Brd4 With 3-(3-(But-3-Yn-1-Yl)-3H-Diazirin-3-Yl)-N-(3-Methyl-[1,2,4]Triazolo[4,3-A]Pyridin-8-Yl)Propanamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-10-21 Classification: TRANSCRIPTION Ligands: R5W, EDO |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5,6-Dichloro-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-06-14 Classification: OXIDOREDUCTASE Ligands: FAD, CL, JHY, GOL |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL, SO4, NA, 54Q, DT |
 |
Organism: Staphylococcus aureus, Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, MN, GOL, NA, EVP |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, NA, GOL, MN, 53M, 53L, 50M |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-12-16 Classification: ISOMERASE |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2015-12-16 Classification: HYDROLASE Ligands: MG, GOL, MFX |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL |
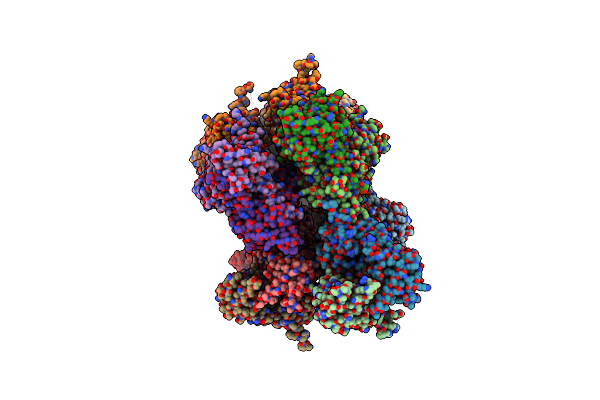 |
Crystal Structure Of The Atpase Region Of Mycobacterium Tuberculosis Gyrb With Amppnp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-09-18 Classification: ISOMERASE Ligands: ANP, MG |
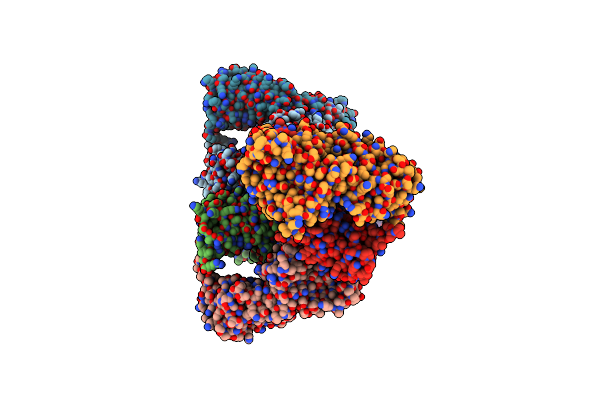 |
Crystal Structure Of The Atpase Region Of Mycobacterium Tuberculosis Gyrb With Amppnp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-09-18 Classification: ISOMERASE Ligands: ANP, MG |
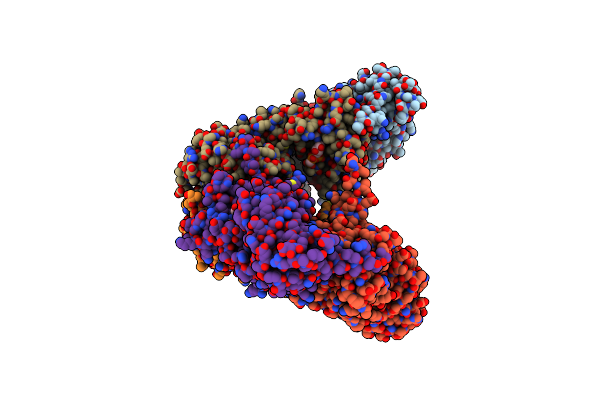 |
Crystal Structure Of The Atpase Region Of Mycobacterium Tuberculosis Gyrb With Amppcp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-09-18 Classification: ISOMERASE Ligands: ACP, MG |
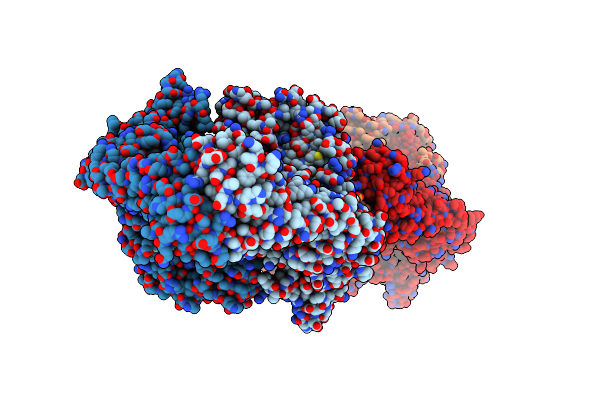 |
The Twinned 3.35A Structure Of S. Aureus Gyrase Complex With Ciprofloxacin And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-08-25 Classification: ISOMERASE/DNA/ANTIBIOTIC Ligands: MN, CPF |
 |
The 3.1A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: CA |
 |
The 2.98A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: HOH |
 |
The 3.5A Crystal Structure Of The Catalytic Core (B'A' Region) Of Staphylococcus Aureus Dna Gyrase Complexed With Gsk299423 And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: RXV |
 |
The 2.1A Crystal Structure Of S. Aureus Gyrase Complex With Gsk299423 And Dna
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-04 Classification: ISOMERASE Ligands: MN, RXV |

