Search Count: 24
 |
Structure Of Pou2F3 Pou Domains Bound To Coactivator Oca-T2 And Dna (2.8 Angstrom Resolution)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION |
 |
Structure Of Pou2F3 Pou Domains Bound To Coactivator Oca-T2 And Dna (2.1 Angstrom Resolution)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION |
 |
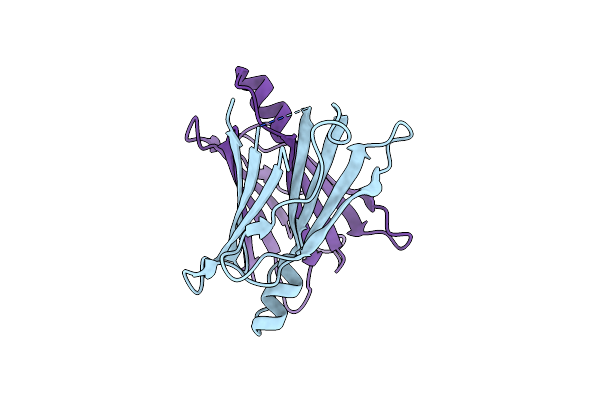
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: BO3 |
 |
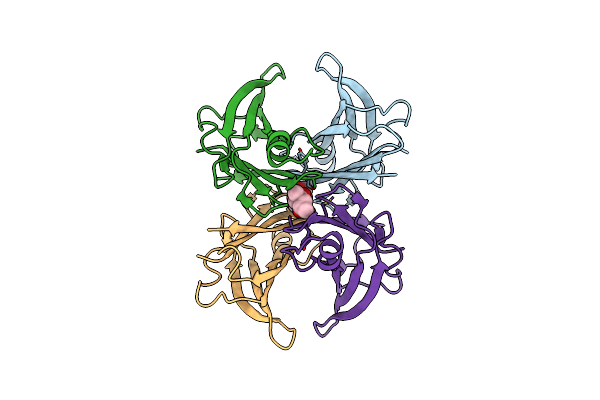
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-17 Classification: PROTEIN FIBRIL |
 |
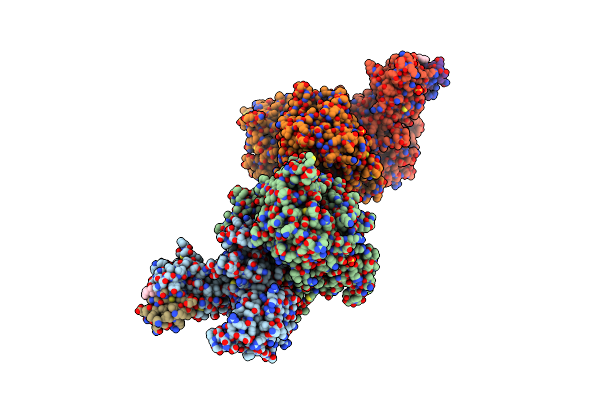
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-01-17 Classification: STRUCTURAL PROTEIN Ligands: MPD |
 |
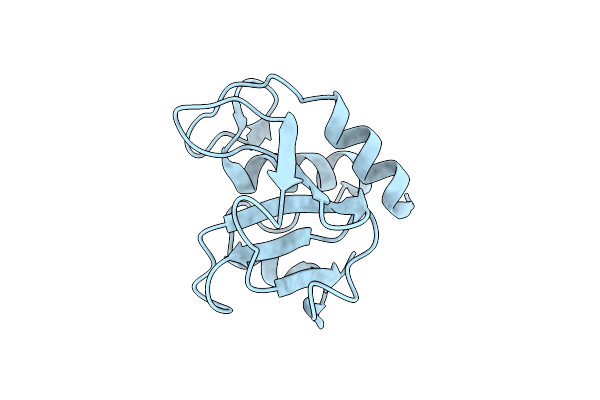
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Ikzf2(Zf2) And The Molecular Glue Dky709
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-03-08 Classification: LIGASE Ligands: ZN, LWK, SO4, EPE |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Ikzf2(Zf2,3) And The Molecular Glue Dky709
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2023-03-08 Classification: LIGASE Ligands: ZN, LWK |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-01-16 Classification: HYDROLASE |
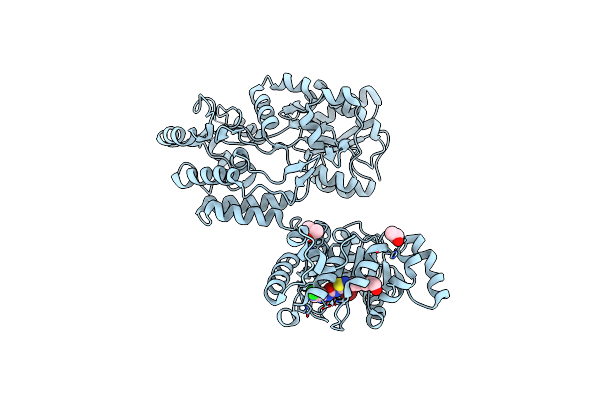 |
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2018-10-31 Classification: Hydrolase/Hydrolase Inhibitor Ligands: J7J, IPA |
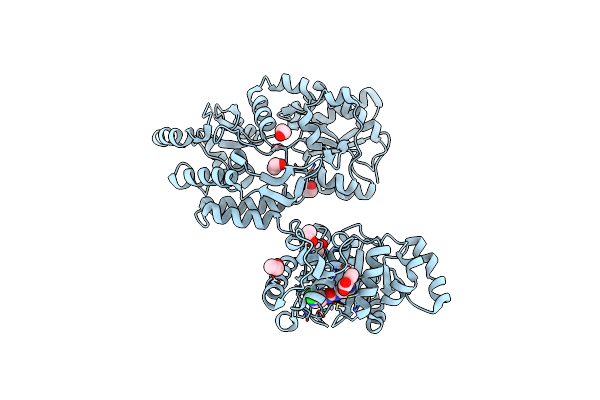 |
Organism: Escherichia coli o157:h7, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2018-10-31 Classification: Hydrolase/Hydrolase Inhibitor Ligands: J7G, EOH |
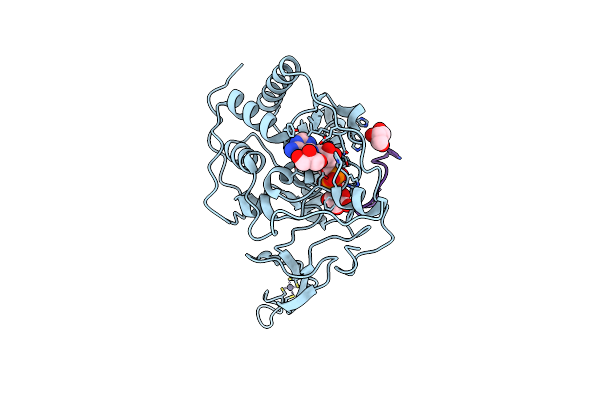 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, APR, GOL |
 |
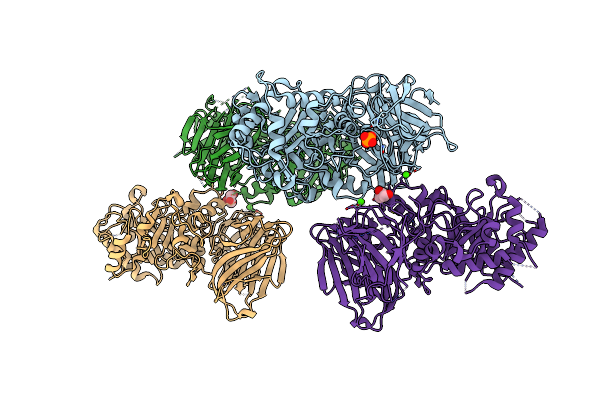
Crystal Structure Of The Nuclease Domain Of Cole7(D493Q Mutant) In Complex With An 18-Bp Duplex Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-11-03 Classification: HYDROLASE/DNA |
 |
Organism: Arthrobacter protophormiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-28 Classification: HYDROLASE Ligands: CA, PO4, GOL, MG |
 |
Glycogen Synthase Kinase-3 Beta In Complex With Bis-(Indole)Maleimide Pyridinophane Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-02-19 Classification: TRANSFERASE Ligands: BIM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-12-27 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-11-22 Classification: HYDROLASE |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-11-22 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-03-01 Classification: CHAPERONE |
 |
Crystal Structure Of The Hspbp1 Core Domain Complexed With The Fragment Of Hsp70 Atpase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-03-01 Classification: CHAPERONE Ligands: AMP |
 |
Crystal Structure Of Bacillus Subtilis Guanine Deaminase. The First Domain-Swapped Structure In The Cytidine Deaminase Superfamily
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2004-07-13 Classification: HYDROLASE Ligands: ZN, IMD |

