Search Count: 29
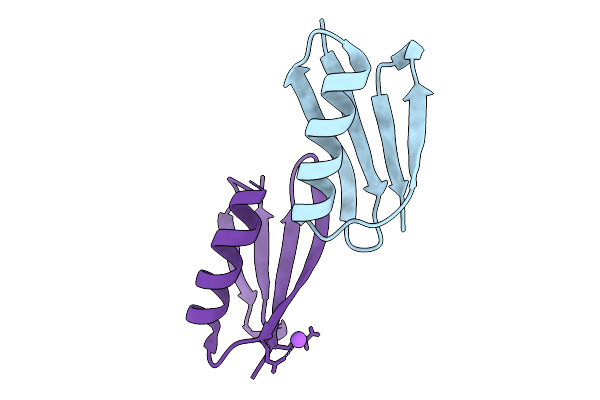 |
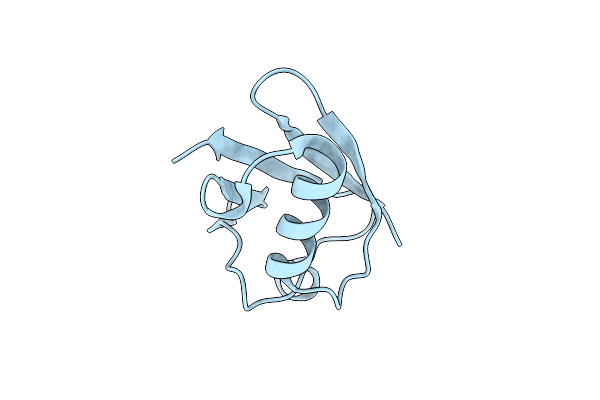
X-Ray Structure Of The B1 Domain Of Streptococcal Protein G Triple Mutant T2Q, N8D, And N37D (Gb1-Qdd).
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: IMMUNE SYSTEM |
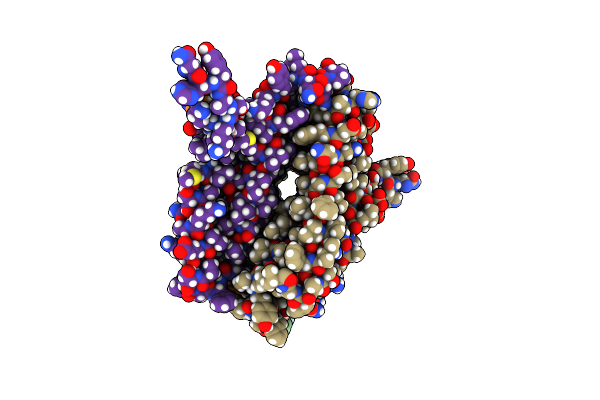 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: MEMBRANE PROTEIN |
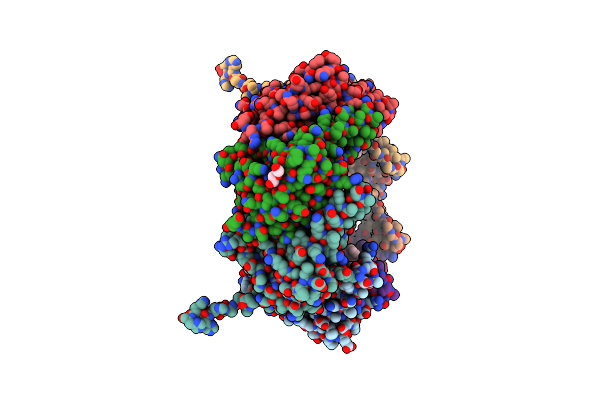 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: BO2, PEG |
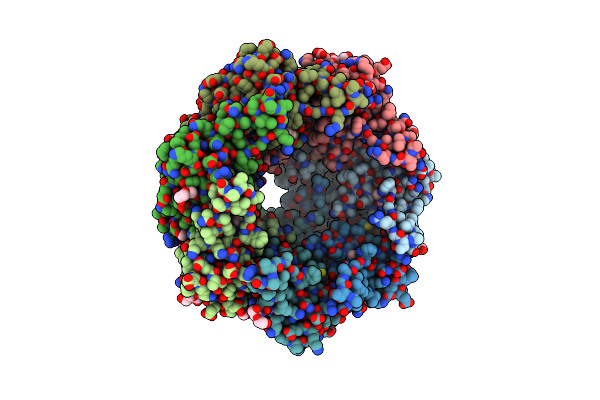 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: PEG |
 |
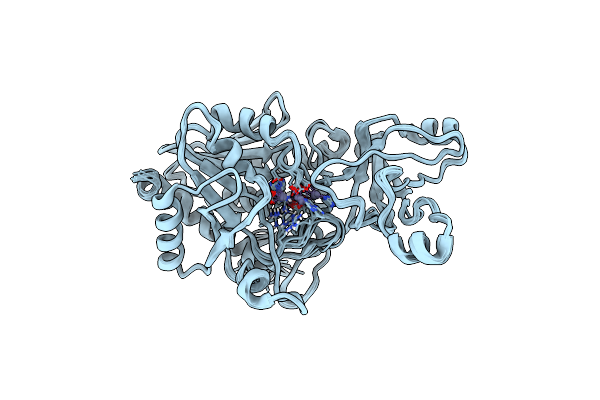
Structure Determination Of The Tetrahedral Aminopeptidase Tet2 From P. Horikoshii By Use Of Combined Solid-State Nmr, Solution-State Nmr And Em Data 4.1 A, Followed By Real_Space_Refinement At 4.1 A
Organism: Pyrococcus horikoshii ot3
Method: ELECTRON MICROSCOPY, SOLUTION NMR Release Date: 2019-08-14 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Combined Solid-State Nmr, Solution-State Nmr And Em Data For Structure Determination Of The Tetrahedral Aminopeptidase Tet2 From P. Horikoshii
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: ELECTRON MICROSCOPY, SOLUTION NMR, SOLID-STATE NMR Release Date: 2018-03-14 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Amino Terminal Domains Of The Nmda Receptor Subunit Glun1 And Glun2B In Complex With Ifenprodil
Organism: Xenopus laevis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2016-03-02 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Amino Terminal Domains Of The Nmda Receptor Subunit Glun1 And Glun2B In Complex With Mk-22
Organism: Xenopus laevis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2016-03-02 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Amino Terminal Domains Of The Nmda Receptor Subunit Glun1 And Glun2B In Complex With Evt-101
Organism: Xenopus laevis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2016-03-02 Classification: TRANSPORT PROTEIN |
 |
Observing The Overall Rocking Motion Of A Protein In A Crystal - Orthorhombic Ubiquitin Crystals Without Zinc.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-10-14 Classification: SIGNALING PROTEIN |
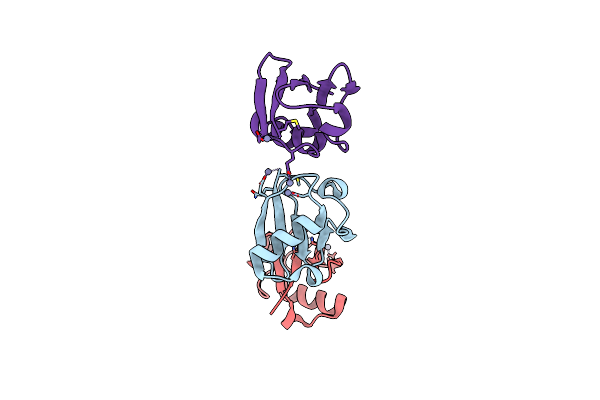 |
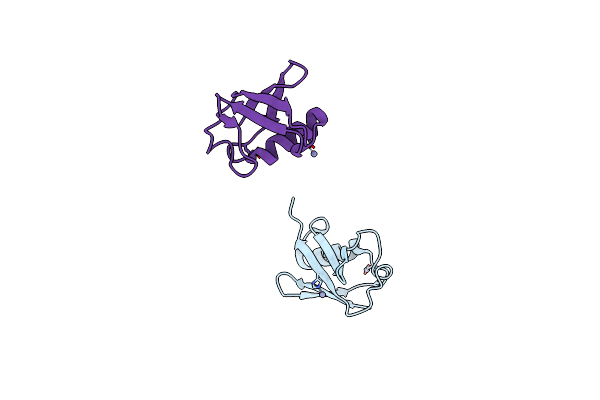
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-10-14 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Observing The Overall Rocking Motion Of A Protein In A Crystal - Cubic Ubiquitin Crystals.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2015-10-14 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Haddock Model Of Bacillus Subtilis L,D-Transpeptidase In Complex With A Peptidoglycan Hexamuropeptide
Organism: Bacillus subtilis
Method: SOLID-STATE NMR Release Date: 2015-01-14 Classification: TRANSFERASE/STRUCTURAL PROTEIN |
 |
 |
Solution Structure Of Copk, A Periplasmic Protein Involved In Copper Resistance In Cupriavidus Metallidurans Ch34
Organism: Cupriavidus metallidurans
Method: SOLUTION NMR Release Date: 2008-05-27 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-01-11 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-03-23 Classification: TRANSPORT PROTEIN Ligands: ZN |
 |
Escherichia Coli Uracil-Dna Glycosylase Complex With Uracil-Dna Glycosylase Inhibitor Protein
Organism: Escherichia coli, Bacillus phage pbs2
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2002-11-10 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2002-11-10 Classification: HYDROLASE |
 |
Escherichia Coli Uracil-Dna Glycosylase Complex With Uracil-Dna Glycosylase Inhibitor Protein
Organism: Escherichia coli, Bacillus phage pbs2
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-11-10 Classification: HYDROLASE/HYDROLASE INHIBITOR |

