Search Count: 62
 |
Vitamin D Receptor Complex With A 5-{4-(M-Carboran-1-Yl)Phenoxy}-4-Hydroxypentanoic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAV |
 |
Vitamin D Receptor Complex With A 4-(M-Carboran-1-Yl)Phenoxyacetic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAW |
 |
Vitamin D Receptor Complex With A 3-{4-(M-Carboran-1-Yl)Phenoxy}Propanoic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAX |
 |
Vitamin D Receptor Complex With A 4-{4-(M-Carboran-1-Yl)Phenoxy}Butanoic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAY |
 |
Vitamin D Receptor Complex With A 5-{4-(M-Carboran-1-Yl)Phenoxy}Pentanoic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAZ |
 |
Vitamin D Receptor Complex With A 4-{4-(M-Carboran-1-Yl)-2-Methylphenoxy}Butanoic Acid Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1MAU |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: A1L81 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: FMT, GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: A1LZO |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: GOL, IPA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: A1L81, ACT |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: A1L81, GOL, PEG |
 |
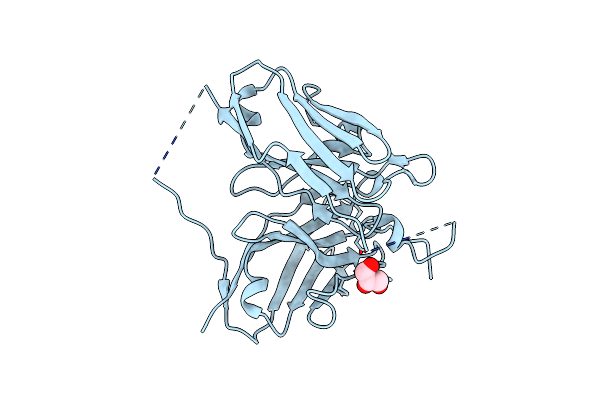
Single-Chain Fv Antibody Of G2 Fused With Antigen Peptide From Chicken Prion Protein
Organism: Gallus gallus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: IMMUNE SYSTEM Ligands: SO4, GOL |
 |
Single-Chain Fv Antibody Of G2 Fused With Antigen Peptide From Chicken Chromosome 6 C10Orf76 Homolog
Organism: Gallus gallus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Vitamin D Receptor Complex With A Dimethylbis((3-Trifluoromethyl)Phenyl)Silane Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION Ligands: A1L7X |
 |
Vitamin D Receptor Complex With A Bis(3-Chlorophenyl)Dimethylsilane Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION Ligands: A1L7Y |
 |
Vitamin D Receptor Complex With A Bis(3-Ethylphenyl)Dimethylsilane Derivative
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION Ligands: A1L7Z |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION Ligands: A1L70 |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSCRIPTION Ligands: A1L71 |

