Search Count: 24
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM/DNA Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM/DNA Ligands: NAG |
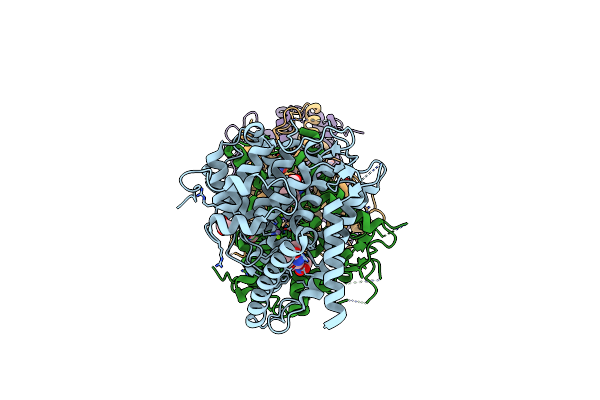 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: X5M, MN, MG, ACT |
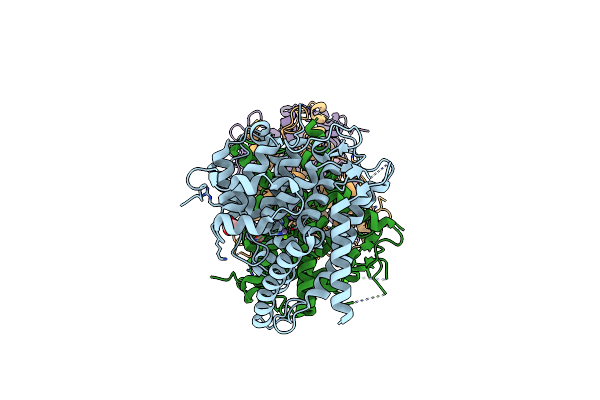 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: MN, MG, ACT, CA |
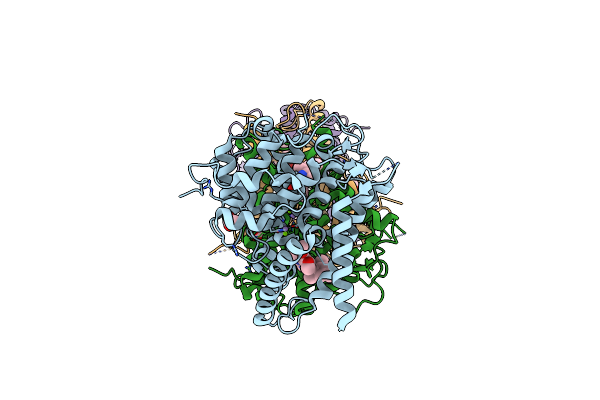 |
Crystal Structure Of The Catalytic Domain Of Human Pde3A Bound To Trequinsin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-06-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: XKG, MN, MG, ACT |
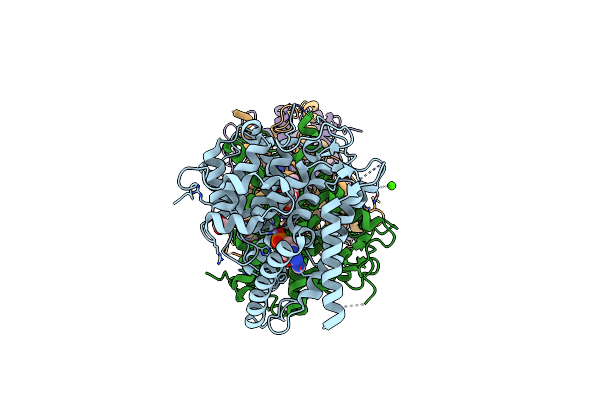 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-06-16 Classification: HYDROLASE Ligands: AMP, MN, MG, CA, ACT |
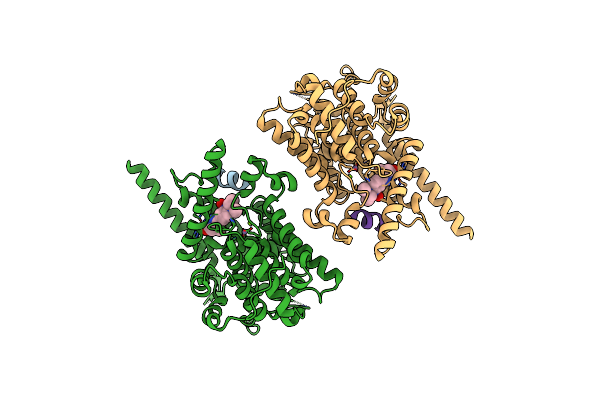 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: HYDROLASE Ligands: MN, MG, X5M |
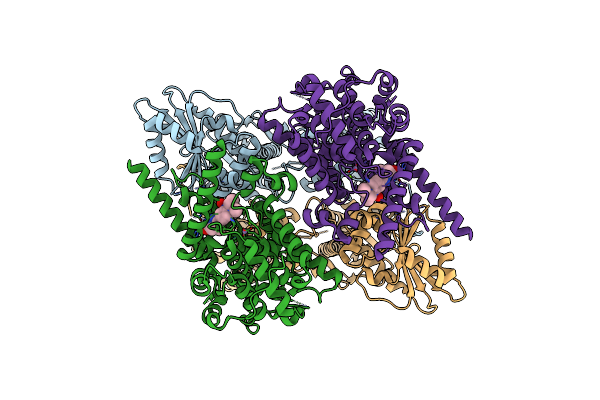 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: RNA BINDING PROTEIN Ligands: ZN, MN, MG, X5M |
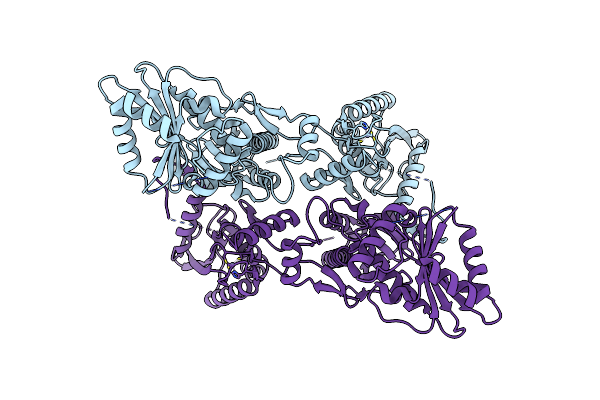 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: RNA BINDING PROTEIN Ligands: ZN |
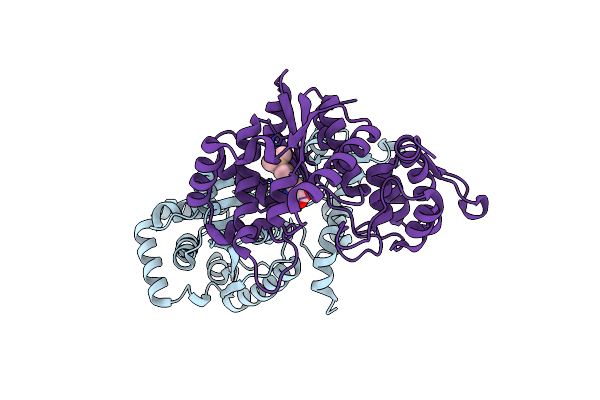 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-03-31 Classification: LIGASE Ligands: TSQ |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-03-31 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: TRANSFERASE/INHIBITOR Ligands: YJG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-11-25 Classification: SPLICING/TRANSFERASE Ligands: QN4, CL, SFG, GOL |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-09-09 Classification: FLUORESCENT PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-08-26 Classification: TRANSFERASE/SPLICING Ligands: SFG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2020-08-26 Classification: Transferase/Splicing Ligands: SFG, ACE |
 |
Crystal Structure Of Rat Sting In Complex With Cyclic Gmp-Amp With 2'5'And 3'5'Phosphodiester Linkage(2'3'-Cgamp)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-10-25 Classification: IMMUNE SYSTEM Ligands: 1SY |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-10-25 Classification: IMMUNE SYSTEM Ligands: SO4 |
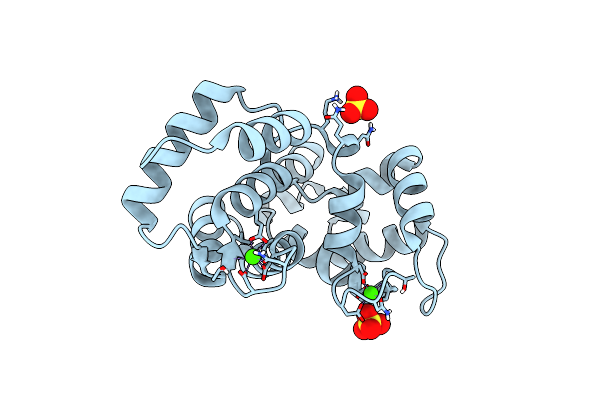 |
Crystal Structure Of Non-Myristoylated E153A Recoverin At 1.2 A Resolution With Calcium Ions Bound To Ef-Hands 2 And 3
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-12-02 Classification: Calcium binding protein Ligands: CA, SO4 |
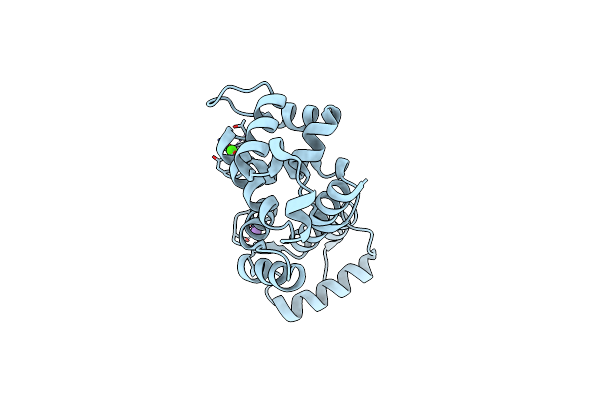 |
Crystal Structure Of Non-Myristoylated E153A Recoverin At 1.35 A Resolution With A Sodium Ion Bound To Ef-Hand 2 And Calcium Ion Bound To Ef-Hand 3
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-12-02 Classification: Calcium Binding protein |

