Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: MPD, SO4 |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: MPD, SO4 |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: A1C3I, SO4, MPD |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: A1C3I, SO4, MPD |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: MPD, A1C3I |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: A1C3I, MPD |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: CO2, A1C3I, MPD |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE/INHIBITOR Ligands: CO2, A1C3I, MPD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: LMN, ZMA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: LMN, ZMA |
 |
Cryo-Em Structure Of The Magnesium Transporter Mgta In An E1-Like Conformation With Bound Mg2+ Ions
Organism: Lactococcus lactis subsp. lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of The Magnesium Transporter Mgta In The E2 Conformation With Bound Lipids And Mg2+
Organism: Lactococcus lactis subsp. lactis cv56
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: METAL TRANSPORT Ligands: MG, CDL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: A1CY9, NA, UNX |
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE |
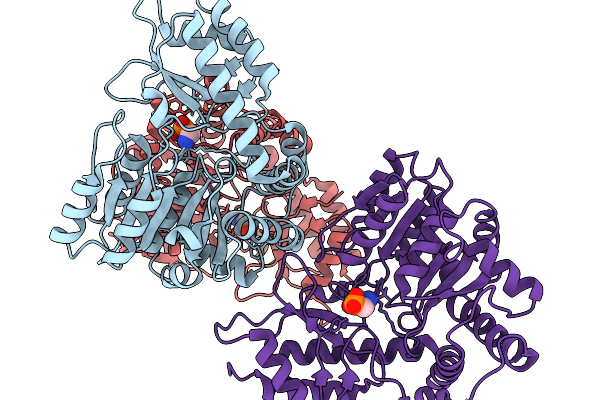 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: P7I |
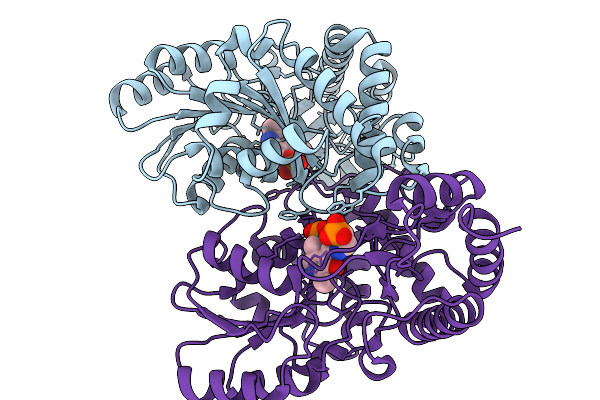 |
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
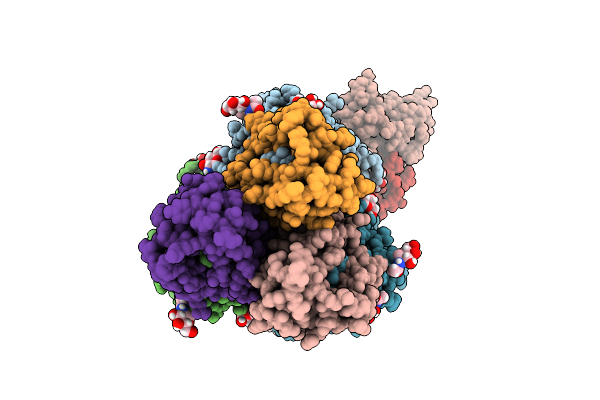 |
Cryo-Em Map Of H5 Ha (A/Jiangsu/Nj210/2023) In Complex With Monoclonal Fab 20D10
Organism: Homo sapiens, Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Map Of H5 Ha (A/Jiangsu/Nj210/2023) In Complex With Monoclonal Fab 12G1
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Map Of H5 Ha (A/Jiangsu/Nj210/2023) In Complex With Monoclonal Fab 6G1
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: ANTITUMOR PROTEIN Ligands: ZN |