Search Count: 571
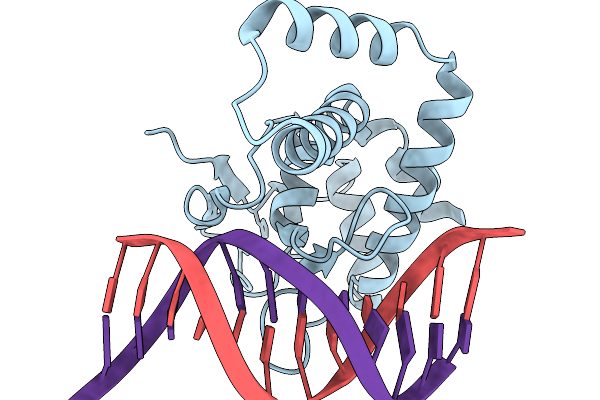 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
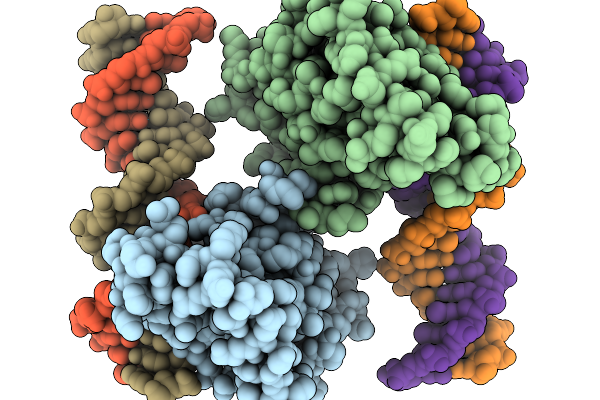 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
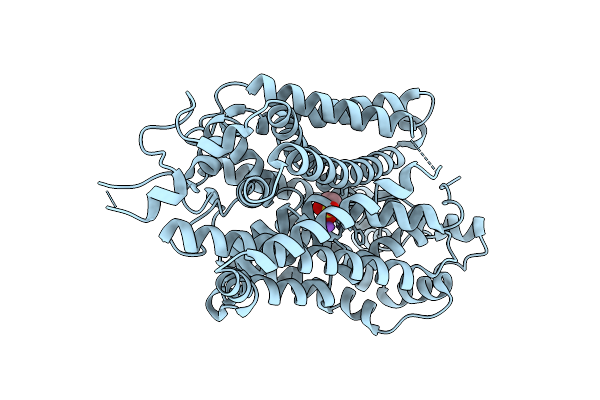
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: TAU, CL, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: BAL, CL, NA |
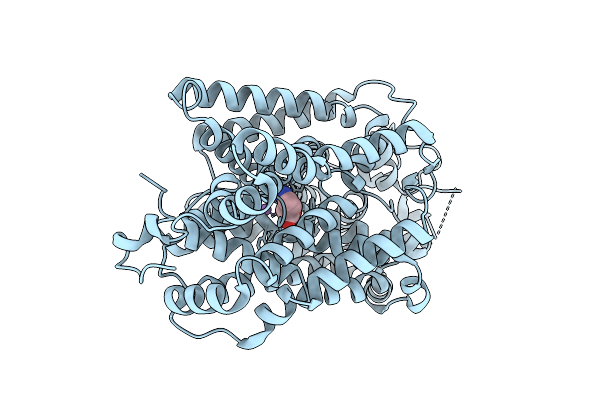 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ABU, CL, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: 3S5, CL |
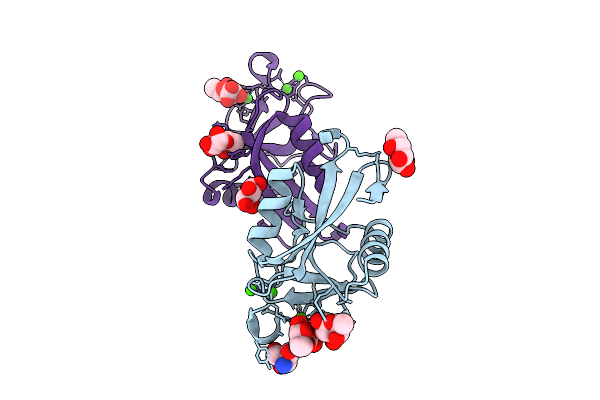 |
Structure Of Collectin-11 (Cl-11) Carbohydrate-Recognition Domain In Complex With L-Fucose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: CA, FUC, FUL, TRS |
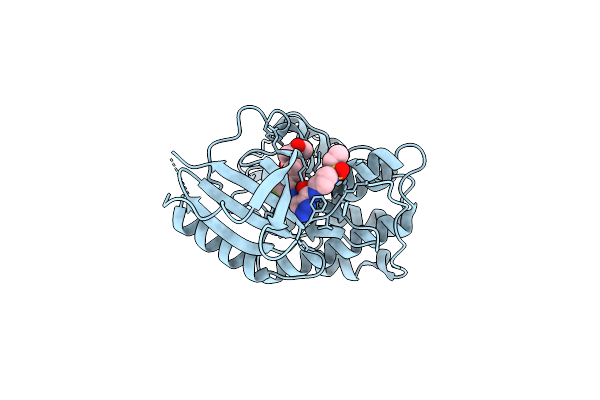 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-09-24 Classification: CELL CYCLE Ligands: A1D6Z |
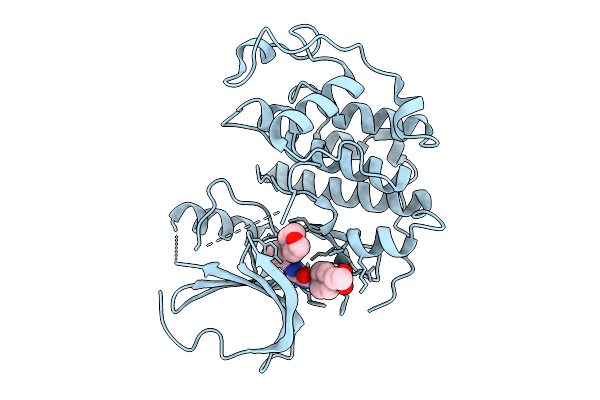 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-09-24 Classification: CELL CYCLE Ligands: A1D60 |
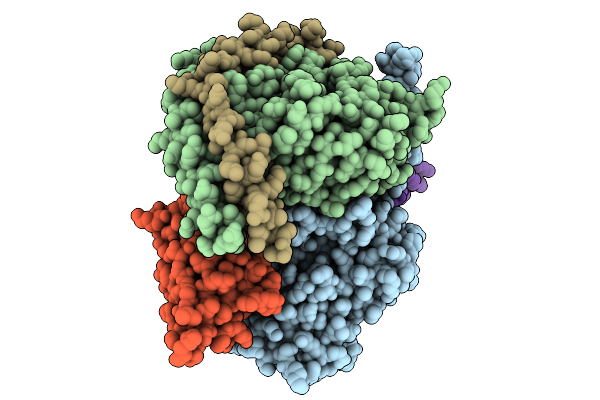 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Crystal Structure Of N-Oxygenase Hrmi With The Diferric Cofactor Partially Loaded
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FE, SO4, GOL |
 |
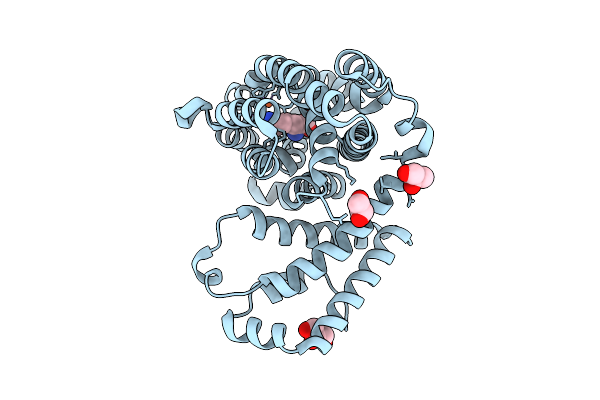
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FE2, SO4, GOL |
 |
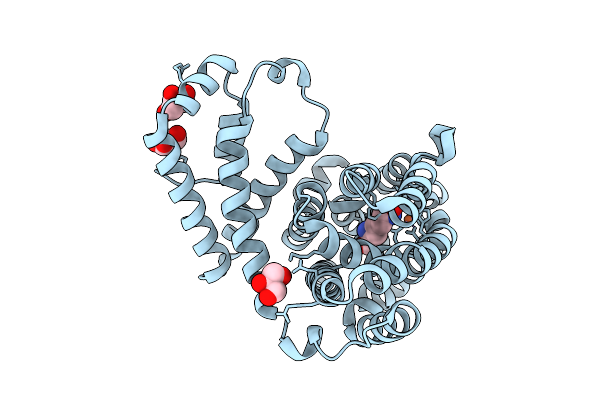
Crystal Structure Of N-Oxygenase Hrmi With The Diferrous Cofactor And Substrate Bound
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FE2, LYS, GOL |
 |
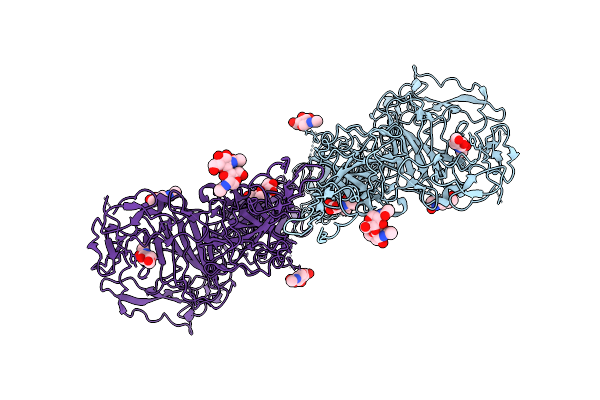
Crystal Structure Of N-Oxygenase Hrmi With The Diferric Cofactor And The N(6)-Hydroxy-L-Lysine Product Bound
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FE, A1BX4, GOL |
 |
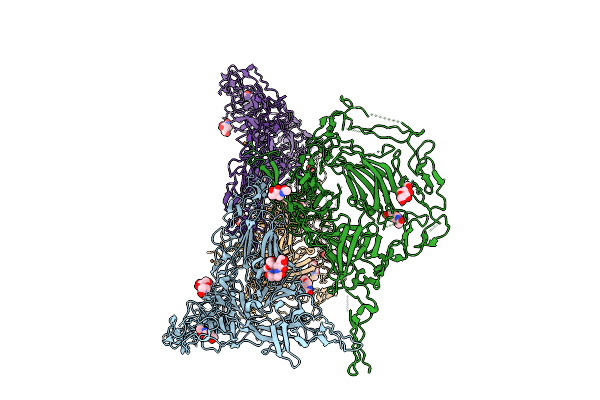
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: NAG, CU |
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |

