Search Count: 1,034
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, A1CG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ONCOPROTEIN/INHIBITOR Ligands: GNP, MG, A1CG4 |
 |
Crystal Structure Of Hras-G12D/Q95H (Gmppnp-Bound) In Complex With Bbo-11818
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ONCOPROTEIN Ligands: GNP, MG, A1CG4 |
 |
Crystal Structure Of A Chimeric Ppm1H Phosphatase With The Flap Domain Of Ppm1J
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: MN |
 |
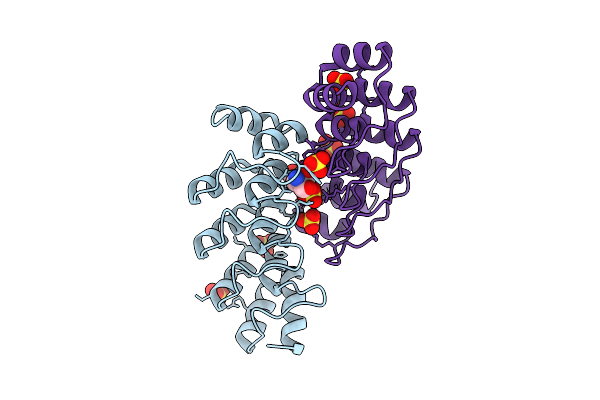
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, FMN, PUJ, GOL |
 |
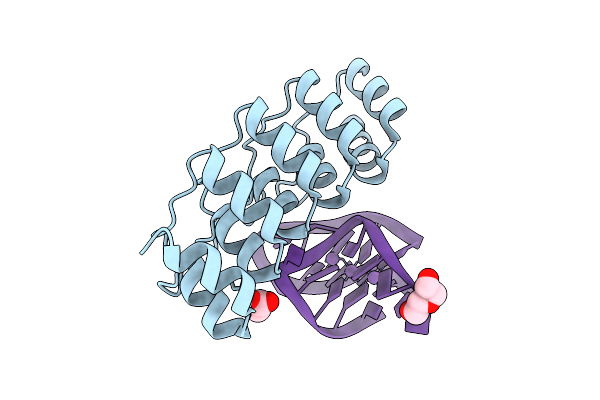
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: SO4, CL, TRS |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: ACY, PEG, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YK, GOL |
 |
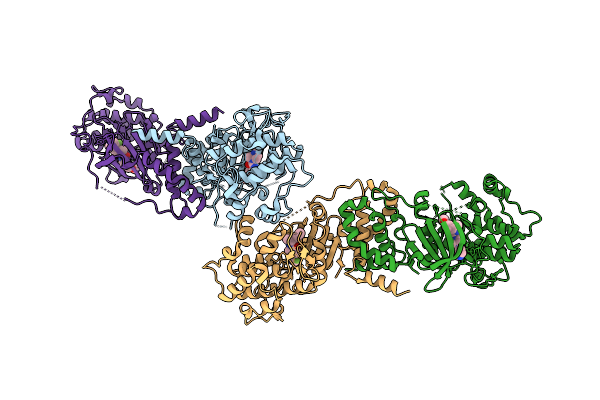
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YV, CL, MES |
 |
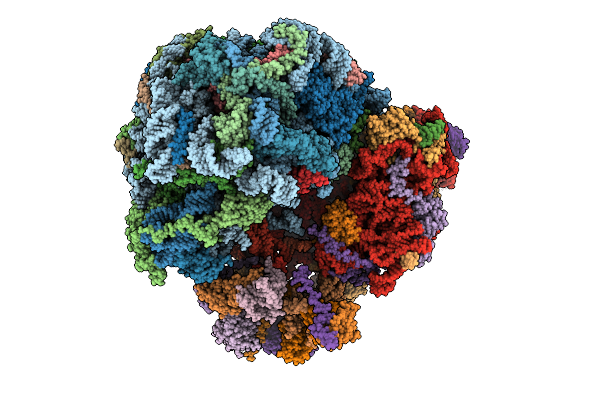
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YO |
 |
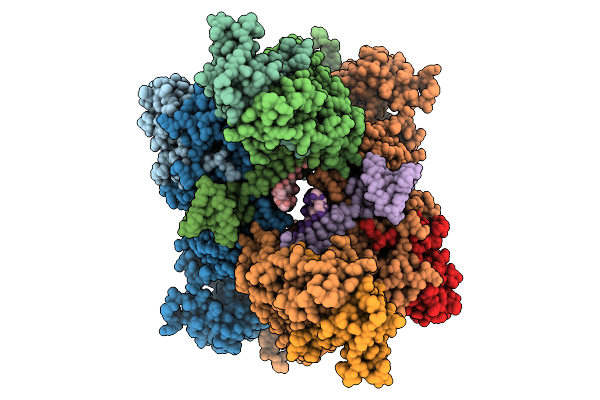
Cryo-Em Structure Of Leishmania Major 80S Ribosome With P/E-Site Trna And Mrna : Lm14Cs1H3 Sko Strain
Organism: Leishmania major strain friedlin
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: RIBOSOME Ligands: K, NA, MG, A1IWA, ZN |
 |
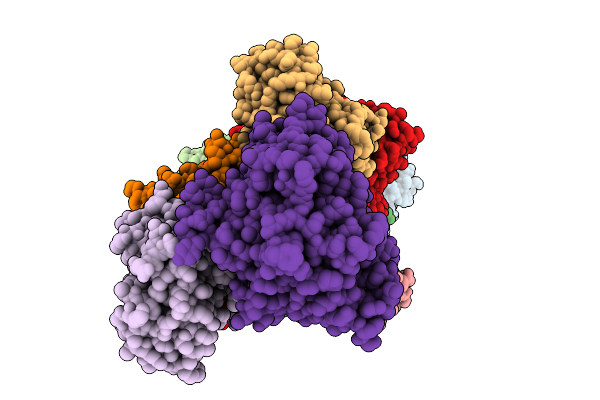
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: ZN, MG |
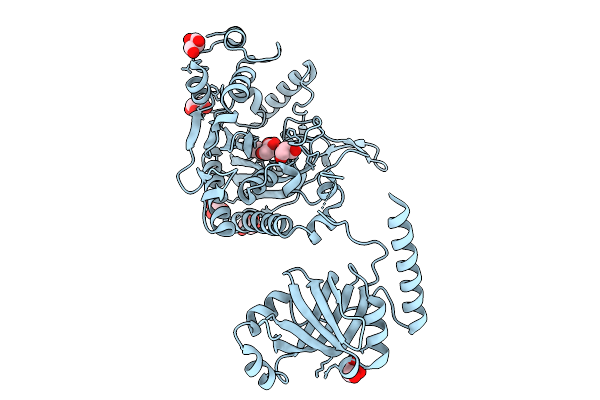 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LIGASE Ligands: LYS, GOL |
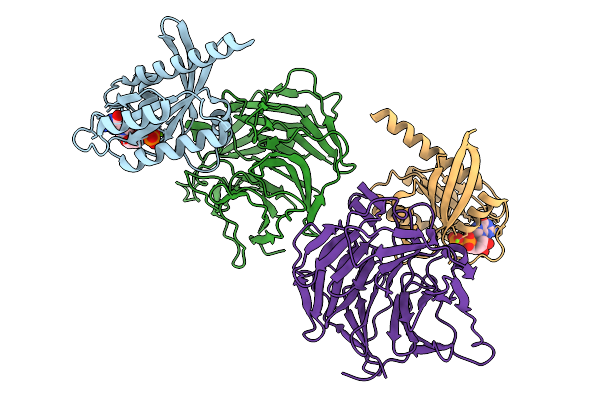 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: GDP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: GDP, MG, FMT, MLA, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: MG, GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: GDP, MG, GOL |
 |
Organism: Porphyromonas gingivalis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: RIBOSOME Ligands: MG, ZN, 62B, K, NA |
 |
Organism: Porphyromonas gingivalis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: RIBOSOME Ligands: MG, K, NA |
 |
Organism: Porphyromonas gingivalis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: RIBOSOME Ligands: MG, ZN, K, NA |

