Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of A Putative Enoyl-Coa Hydratase/Isomerase Family Protein From Hyphomonas Neptunium
Organism: Hyphomonas neptunium (strain atcc 15444)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-10-14 Classification: ISOMERASE Ligands: PG4, MLT |
 |
Crystal Structure Of A Putative Enoyl-Coa Hydratase/Isomerase From Novosphingobium Aromaticivorans
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2014-12-03 Classification: ISOMERASE |
 |
Crystal Structure Of A Purine Nucleoside Phosphorylase (Psi-Nysgrc-029736) From Agrobacterium Vitis
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: HPA |
 |
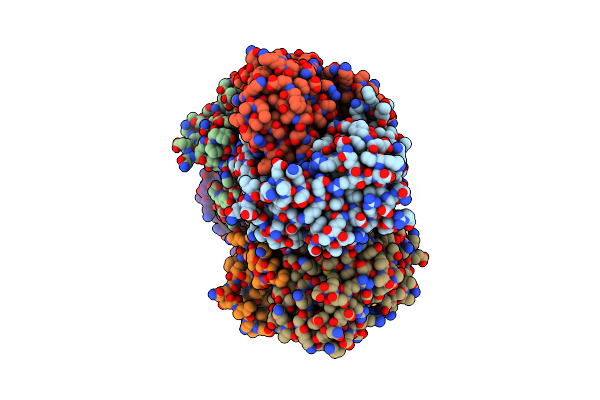
Crystal Structure Of Putative Polyketide Cyclase (Protein Sma1630) From Sinorhizobium Meliloti At 2.3 A Resolution
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-09-10 Classification: TRANSFERASE |
 |
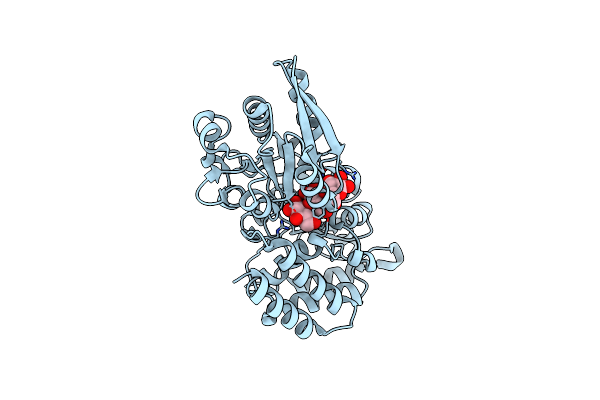
Crystal Structure Of A Putative Uridine Phosphorylase From Actinobacillus Succinogenes 130Z (Target Nysgrc-029667 )
Organism: Actinobacillus succinogenes
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-27 Classification: TRANSFERASE Ligands: GOL |
 |
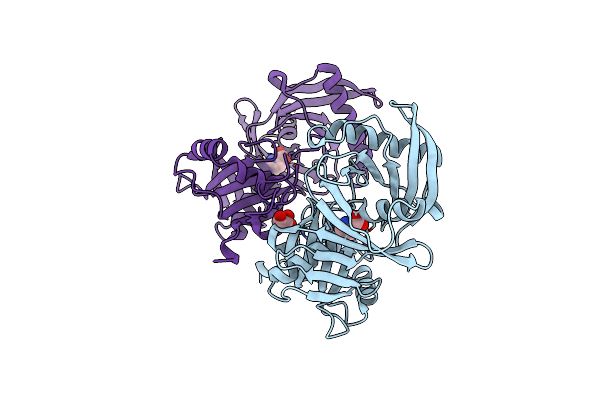
Crystal Structure Of Sugar Transporter Atu4361 From Agrobacterium Fabrum C58, Target Efi-510558, With Bound Maltotriose
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2014-07-16 Classification: TRANSPORT PROTEIN |
 |
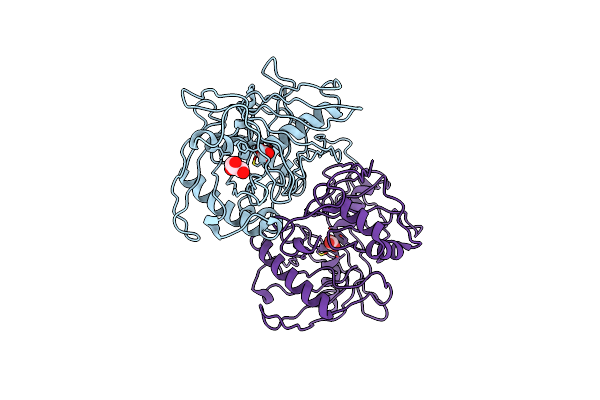
Crystal Structure Of A 4-Hydroxyproline Epimerase From Burkholderia Multivorans Atcc 17616, Target Efi-506586, Open Form, With Bound Pyrrole-2-Carboxylate
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-07 Classification: ISOMERASE Ligands: GOL, PYC |
 |
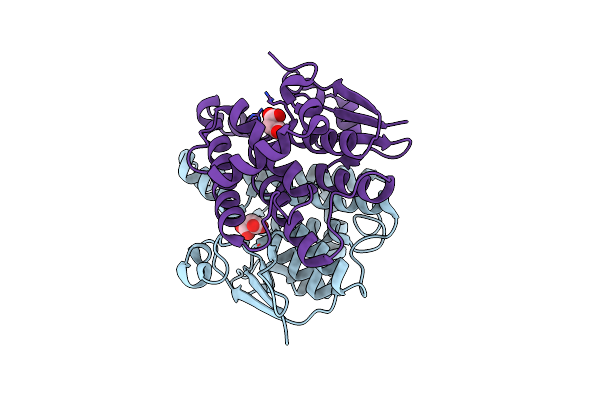
Crystal Structure Of Probable Proline Racemase From Agrobacterium Radiobacter K84, Target Efi-506561, With Bound Carbonate
Organism: Agrobacterium radiobacter k84
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-30 Classification: ISOMERASE Ligands: BCT, GOL |
 |
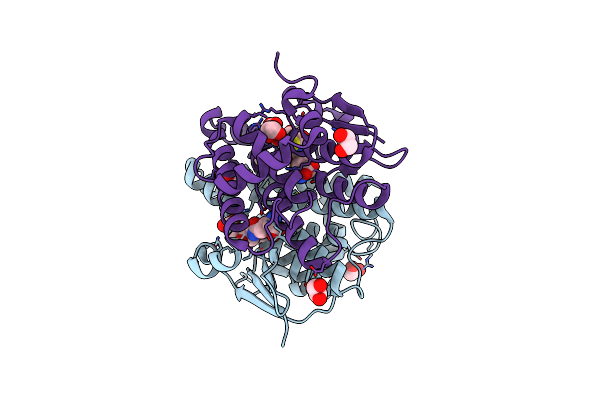
Crystal Structure Of Maleylacetoacetate Isomerase From Methylobacteriu Extorquens Am1 With Bound Malonate (Target Efi-507068)
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-04-09 Classification: TRANSFERASE Ligands: MLA, CL |
 |
Crystal Structure Of Maleylacetoacetate Isomerase From Methylobacteriu Extorquens Am1 With Bound Malonate And Gsh (Target Efi-507068)
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-09 Classification: TRANSFERASE Ligands: MLA, GSH, EDO |
 |
Crystal Structure Of The Cbs Pair Of A Putative D-Arabinose 5-Phosphate Isomerase From Methylococcus Capsulatus In Complex With Cmp-Kdo
Organism: Methylococcus capsulatus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-01-22 Classification: ISOMERASE Ligands: CMK, GOL |
 |
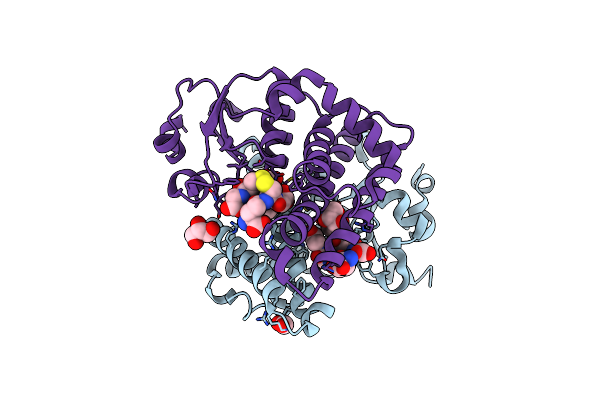
Crystal Structure Of Glutathione Transferase Smc00097 From Sinorhizobium Meliloti, Target Efi-507275, With One Glutathione Bound Per One Protein Subunit
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-20 Classification: TRANSFERASE Ligands: GSH |
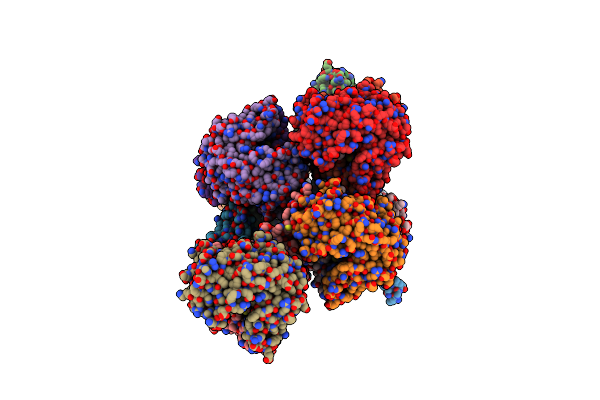 |
Crystal Structure Of Glutathione Transferase Bbta-3750 From Bradyrhizobium Sp., Target Efi-507290, With One Glutathione Bound
Organism: Bradyrhizobium sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-20 Classification: TRANSFERASE Ligands: GSH |
 |
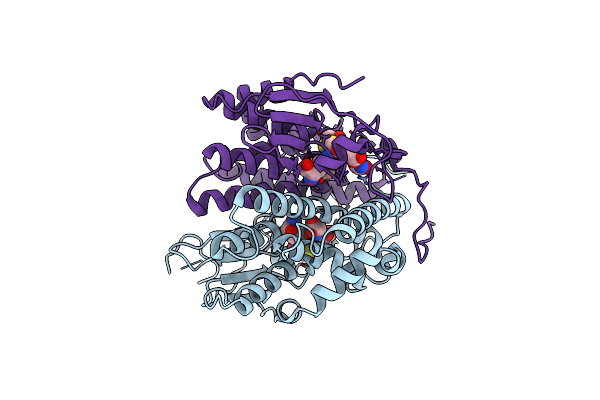
Crystal Structure Of Glutathione Transferase Pput_1760 From Pseudomonas Putida, Target Efi-507288, With One Glutathione Disulfide Bound Per One Protein Subunit
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-11-06 Classification: TRANSFERASE Ligands: GDS, GOL, FMT |
 |
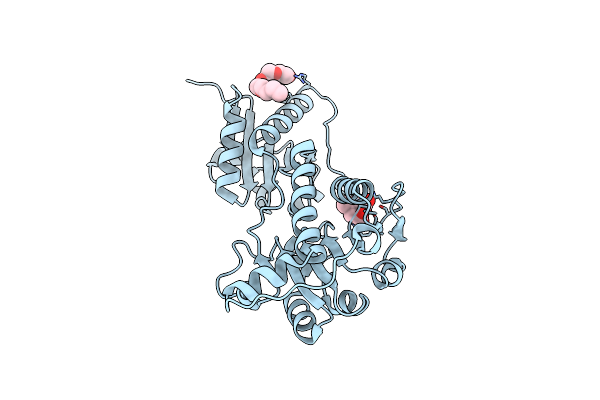
Crystal Structure Of Nu-Class Glutathione Transferase Yghu From Streptococcus Sanguinis Sk36, Complex With Glutathione Disulfide, Target Efi-507286
Organism: Streptococcus sanguinis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-10-16 Classification: TRANSFERASE Ligands: GDS, ACT |
 |
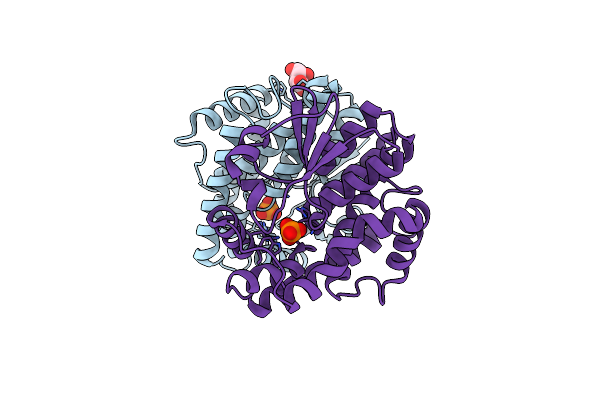
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase Family Protein From Klebsiella Pneumoniae 342
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: PE4, CIT |
 |
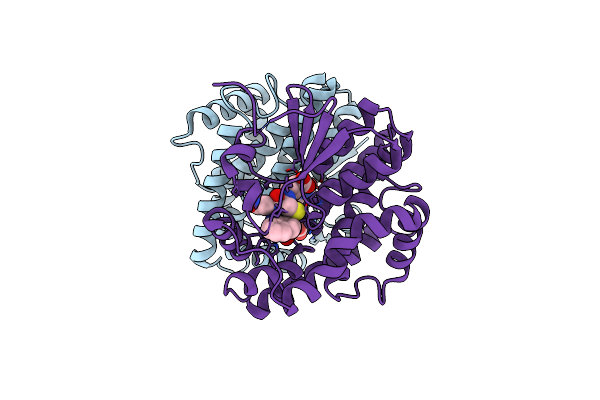
Crystal Structure Of Human Glutathione Transferase Theta-2, Complex With Inorganic Phosphate, Gsh Free, Target Efi-507257
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-09-25 Classification: TRANSFERASE Ligands: TLA, PO4 |
 |
Crystal Structure Of Human Glutathione Transferase Theta-2, Complex With Glutathione And Unknown Ligand, Target Efi-507257
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-09-25 Classification: TRANSFERASE Ligands: GSH, UNL, FMT |
 |
Crystal Structure Of Glutathione S-Transferase From Sinorhizobium Meliloti 1021, Nysgrc Target 021389
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Glutathione Transferase Bgramdraft_1843 From Burkholderia Graminis, Target Efi-507289, With Traces Of One Gsh Bound
Organism: Burkholderia graminis
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: FMT, GSH |