Search Count: 13
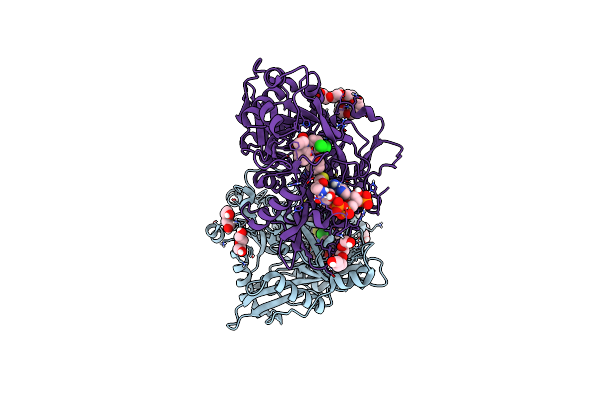 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 1
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOF, PEG, CL, PG4, GOL |
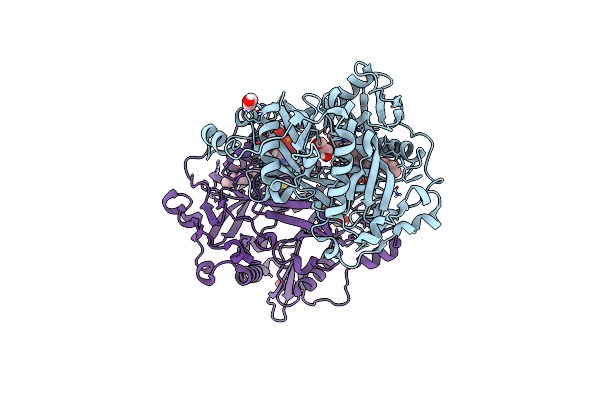 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: MYA, EDO, PEG, GOL |
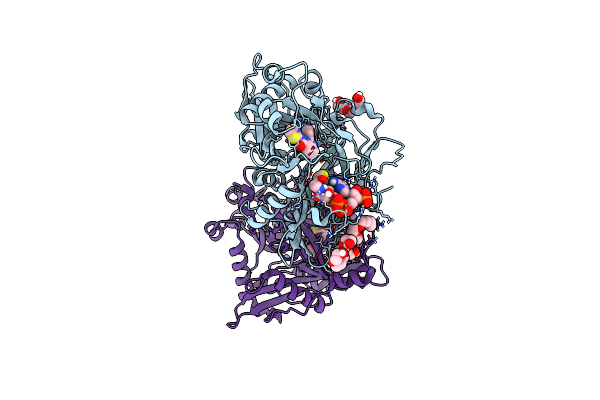 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Compound-2
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-31 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, XOL, PG4, 1PE, PGE, EDO, CL |
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-11 Classification: PROTEIN BINDING Ligands: PGE, EDO, PEG |
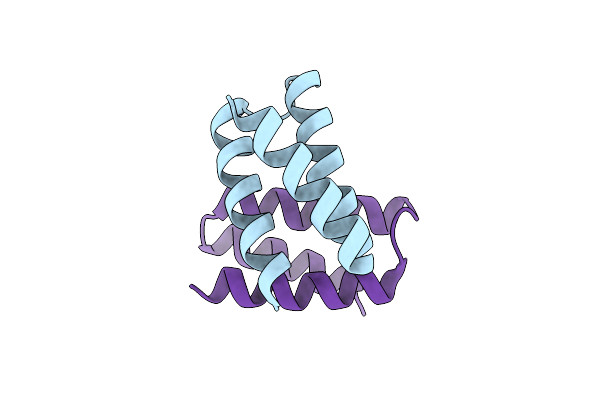 |
Unbound State Of A De Novo Designed Protein Binder To The Human Interleukin-7 Receptor
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-11 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The De Novo Designed Binding Protein H3Mb In Complex With The 1968 Influenza A Virus Hemagglutinin
Organism: Influenza a virus (strain a/hong kong/1/1968 h3n2), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/DE NOVO PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-04-20 Classification: DE NOVO PROTEIN Ligands: NAG, EDO, SO4 |
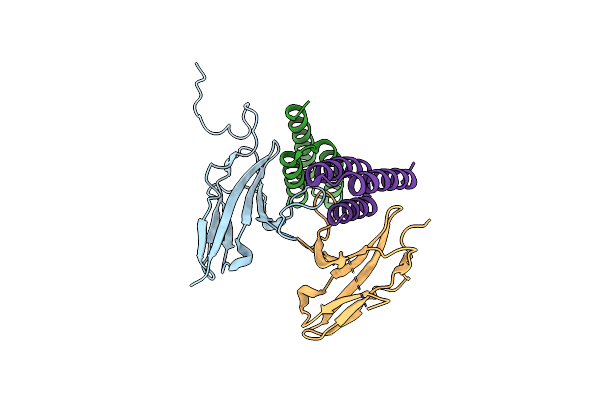 |
Crystal Structure Of Fgfr4 Domain 3 In Complex With A De Novo-Designed Mini-Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-04-06 Classification: SIGNALING PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2022-04-06 Classification: DE NOVO PROTEIN |
 |
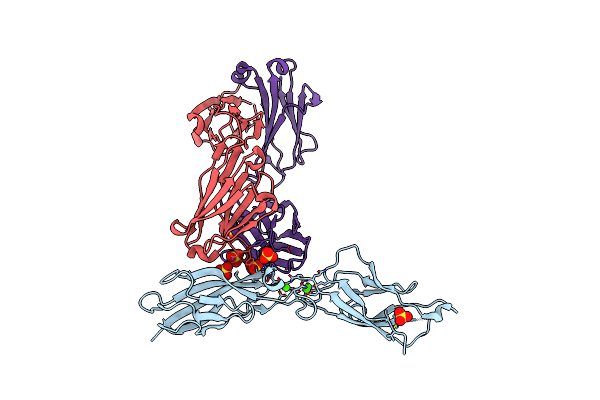
Crystal Structure Of Human E-Cadherin Ec1-5 Bound By Mouse Monoclonal Antibody Fab Mab-1_19A11
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-01-26 Classification: cell adhesion/immune system Ligands: EDO, NAG, PG4, MAN, BMA, CA |
 |
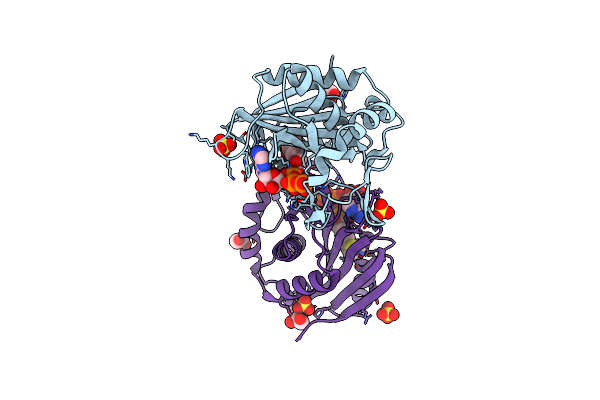
Crystal Structure Of Human E-Cadherin Bound By Mouse Monoclonal Antibody 66E8Fab
Organism: Mus musculus, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2020-01-29 Classification: CELL ADHESION/Immune System Ligands: SO4, CA |
 |
Crystal Structure Of Biotin Acetyl Coenzyme A Carboxylase Synthetase From Helicobacter Pylori With Bound Biotinylated Atp
Organism: Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-04-04 Classification: LIGASE Ligands: F5D, SO4, EDO |
 |
Crystal Structure Of Glycylpeptide N-Tetradecanoyltransferase From Plasmodium Vivax In Complex With Inhibitor Imp-0001088
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-03-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MYA, KFK, SO4, CL |

