Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
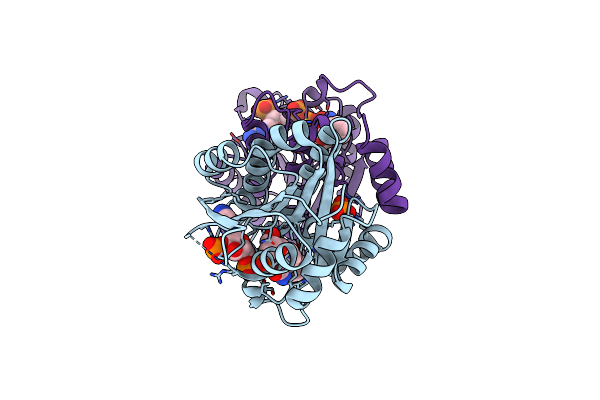 |
Crystal Structure Of A Short Chain Dehydrogenase From Burkholderia Cenocepacia J2315 In Complex With Nadp.
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: NAP, BEZ, PO4 |
 |
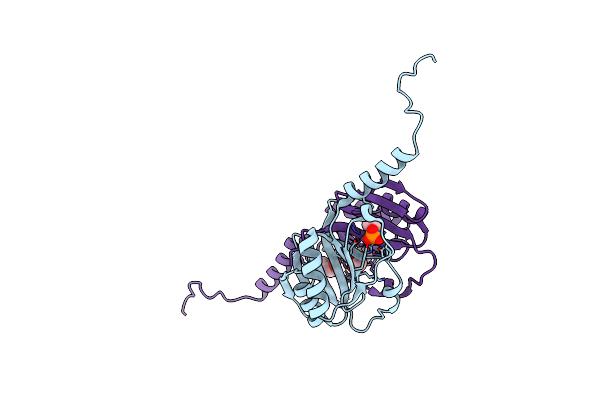
Crystal Structure Of S.Aureus Glyceraldehyde-3-Phosphate-Dehydrogenase (Gap) Containing Oxidized Cys151
Organism: Staphylococcus aureus (strain mrsa252)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-12-28 Classification: OXIDOREDUCTASE |
 |
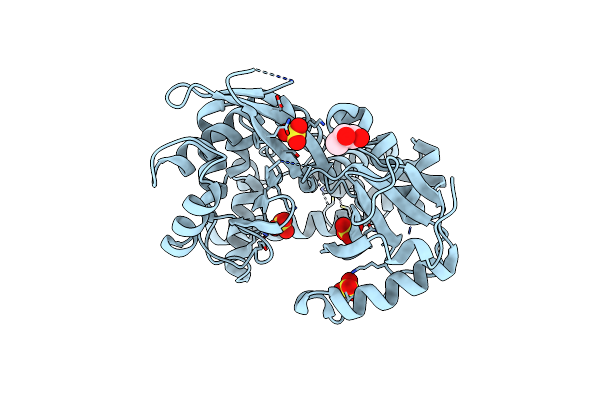
Crystal Structure Of Metallothiol Transferase Fosb 2 From Bacillus Anthracis Str. Ames
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-03-20 Classification: TRANSFERASE Ligands: ZN, FCN, PGE |
 |
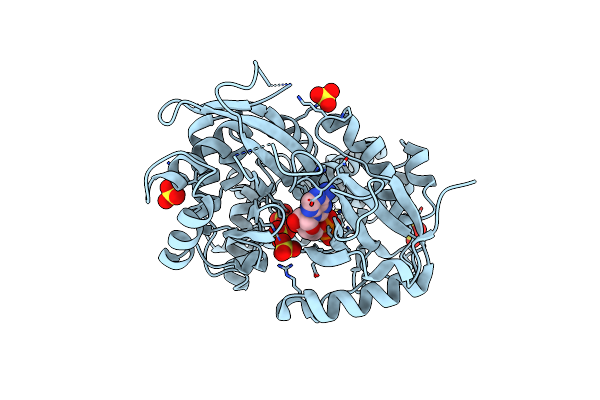
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: ZN, SO4, GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: ANP, ZN, SO4, CIT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: ADP, ZN, SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: ZN, ANP, 4MY |
 |
Crystal Structure Of Pseudouridine Synthase Rlua: Indirect Sequence Readout Through Protein-Induced Rna Structure
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-11-21 Classification: Lyase/RNA Ligands: FOU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-08-09 Classification: LYASE/RNA Ligands: SO4 |