Search Count: 7
 |
Crystal Structure Of An Enoyl-[Acyl-Carrier-Protein] Reductase Inha From Mycobacterium Fortuitum Bound To Nad And Nitd-916
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: NAD, 3KY, NA, CL |
 |
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: 365, SO4 |
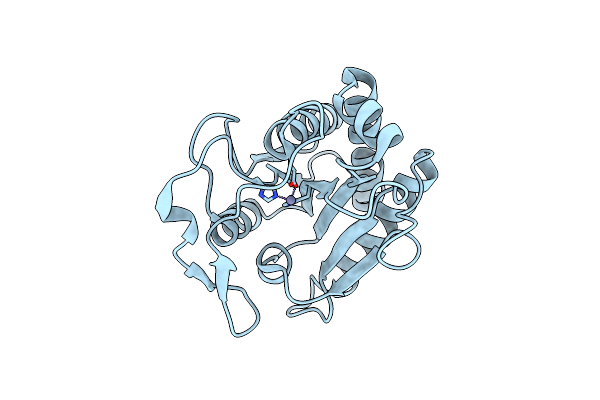 |
Crystal Structure Of The Apo Form Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus.
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
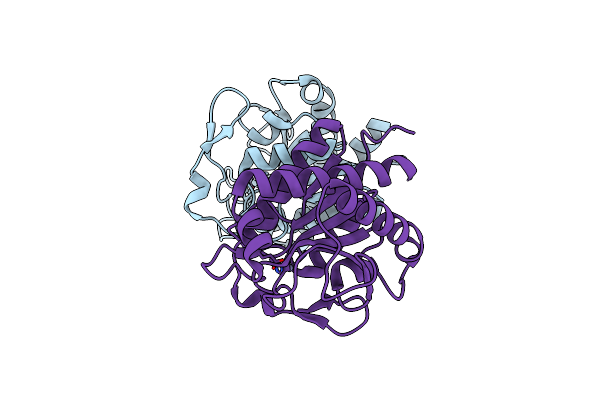 |
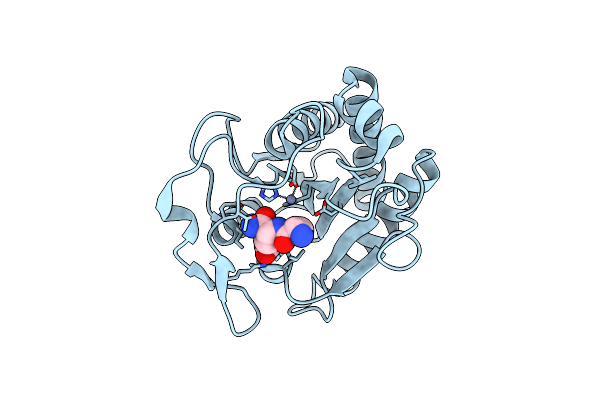
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
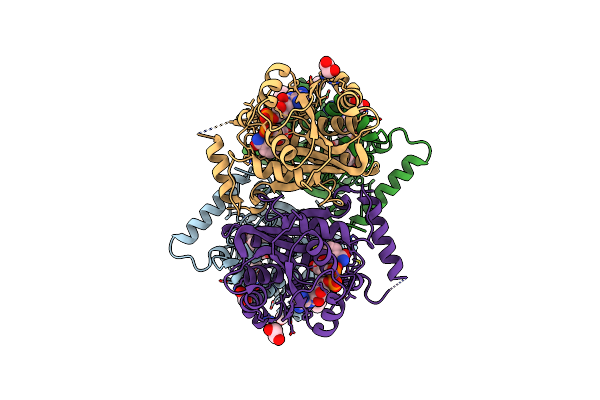
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus Bound To L-Alanine-D-Isoglutamine
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZGL, ALA, ZN |
 |
Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase [Nadh] From Mycobacterium Fortuitum Bound To Nad
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, ACT |

