Search Count: 15
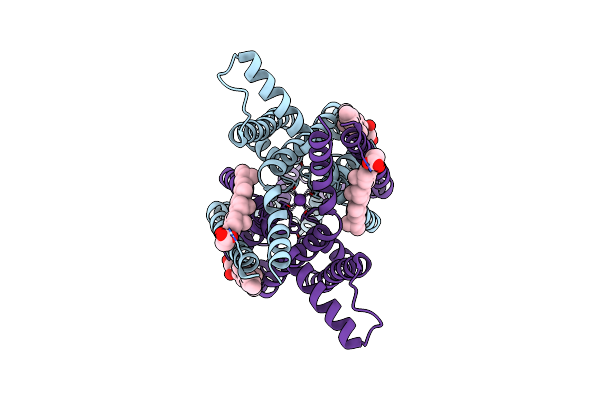 |
Structure Of The Human Two Pore Domain Potassium Ion Channel Task-3 (K2P9.1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: Y01, K |
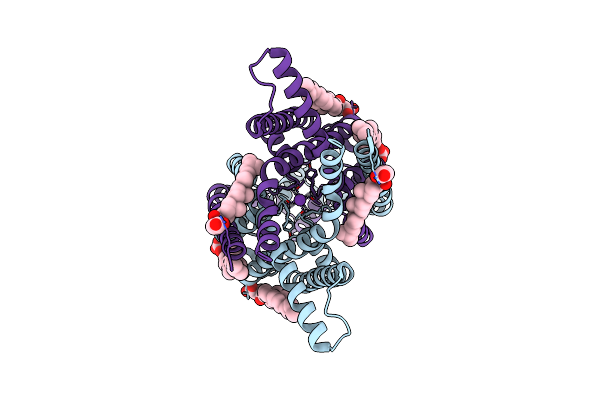 |
Structure Of The Human Two Pore Domain Potassium Ion Channel Task-3 (K2P9.1) G236R Mutant
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: Y01, K |
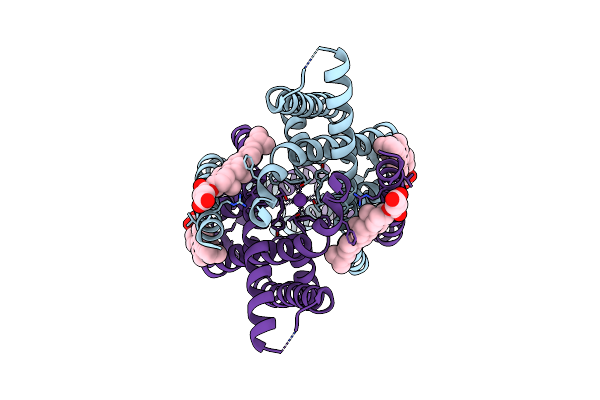 |
Structure Of The Human Two Pore Domain Potassium Ion Channel Task-1 (K2P3.1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: MEMBRANE PROTEIN Ligands: Y01, K |
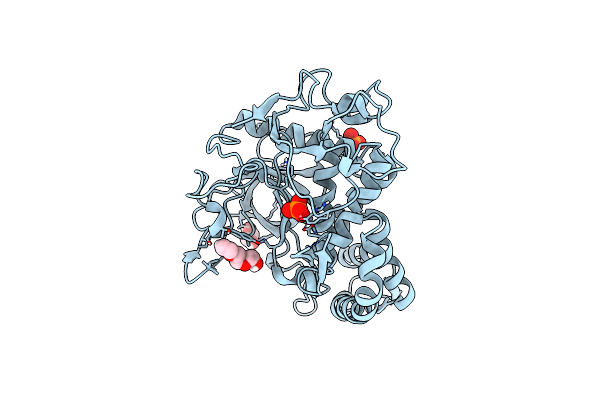 |
Crystal Structure Of The Catalytic Domain Of A Botulinum Neurotoxin Homologue From Enterococcus Faecium
Organism: Enterococcus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-30 Classification: TOXIN Ligands: ZN, PO4, PGE, EDO |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-08-02 Classification: IMMUNE SYSTEM Ligands: NAG, GLC, SO4, BME |
 |
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: CO |
 |
Propionibacterium Shermanii Transcarboxylase 5S Subunit Bound To Oxaloacetate
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: OAA, CO |
 |
Propionibacterium Shermanii Transcarboxylase 5S Subunit Bound To Pyruvic Acid
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: CO, PYR |
 |
Propionibacterium Shermanii Transcarboxylase 5S Subunit Bound To 2-Ketobutyric Acid
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: CO, 2KT |
 |
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: CO |
 |
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-09-07 Classification: TRANSFERASE Ligands: CO |
 |
Crystal Structure Of An Insect Delta-Class Glutathione S-Transferase From A Ddt-Resistant Strain Of The Malaria Vector Anopheles Gambiae
Organism: Anopheles gambiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-12-09 Classification: TRANSFERASE Ligands: GTX |
 |
Transcarboxylase 12S Crystal Structure: Hexamer Assembly And Substrate Binding To A Multienzyme Core (With Methylmalonyl-Coenzyme A And Methylmalonic Acid Bound)
Organism: Propionibacterium freudenreichii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-05-20 Classification: TRANSFERASE Ligands: CD, MCA, DXX, MPD |
 |
Transcarboxylase 12S Crystal Structure: Hexamer Assembly And Substrate Binding To A Multienzyme Core (With Hydrolyzed Methylmalonyl-Coenzyme A Bound)
Organism: Propionibacterium freudenreichii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-05-20 Classification: TRANSFERASE Ligands: CD, MCA, MPD |

