Search Count: 8
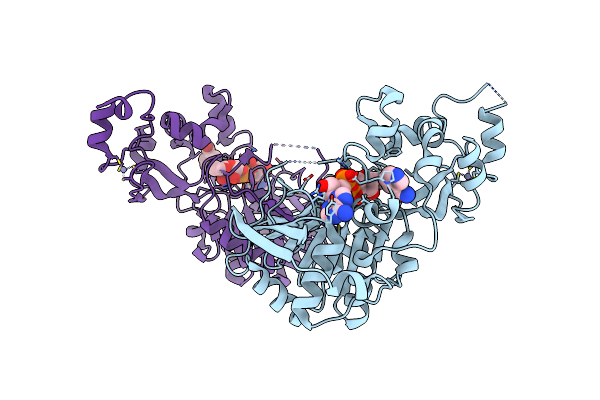 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-04-08 Classification: HYDROLASE Ligands: ZN, XYQ, NCA |
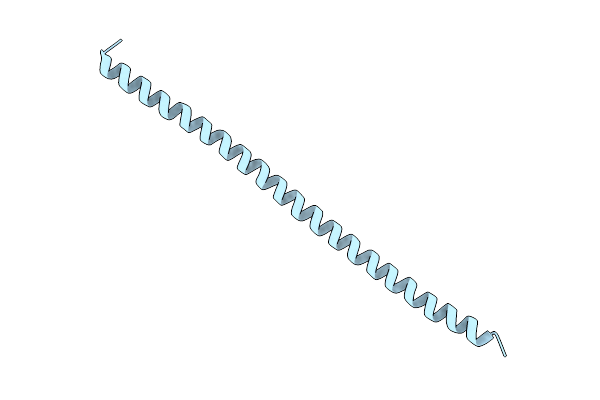 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-06-24 Classification: Transcription repressor |
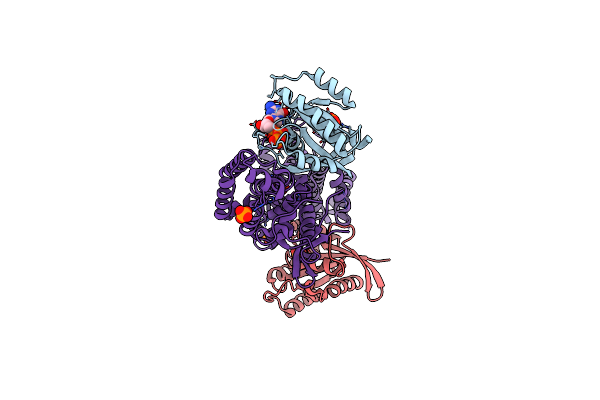 |
Structural Evidence For Feedback Activation By Rasgtp Of The Ras-Specific Nucleotide Exchange Factor Sos
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-04-01 Classification: SIGNALING PROTEIN Ligands: MG, PO4, GTP |
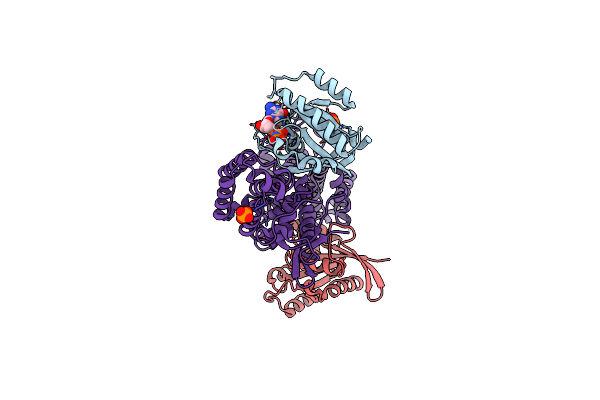 |
Structural Evidence For Feedback Activation By Rasgtp Of The Ras-Specific Nucleotide Exchange Factor Sos
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2003-04-01 Classification: SIGNALING PROTEIN Ligands: MG, PO4, GNP |
 |
Structural Evidence For Feedback Activation By Rasgtp Of The Ras-Specific Nucleotide Exchange Factor Sos
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-04-01 Classification: SIGNALING PROTEIN Ligands: MG, PO4, GNP |
 |
Structural Evidence For Feedback Activation By Rasgtp Of The Ras-Specific Nucleotide Exchange Factor Sos
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2003-04-01 Classification: SIGNALING PROTEIN Ligands: MG, GTP, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2002-11-06 Classification: SIGNALING PROTEIN Ligands: MG, CA, GNP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2002-11-06 Classification: SIGNALING PROTEIN Ligands: MG, GDP |

