Search Count: 11
All
Selected
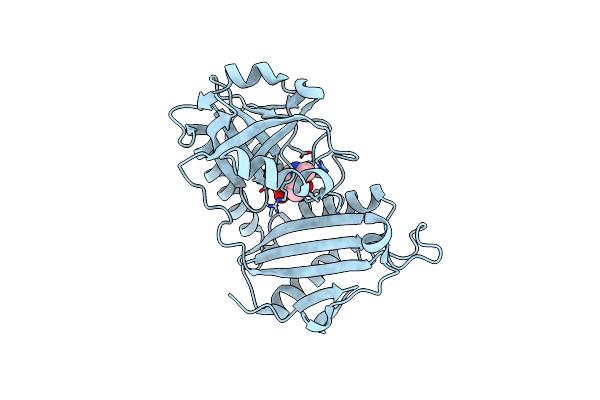 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis In The Holo Form Obtained At Ph 7.0
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: PLP |
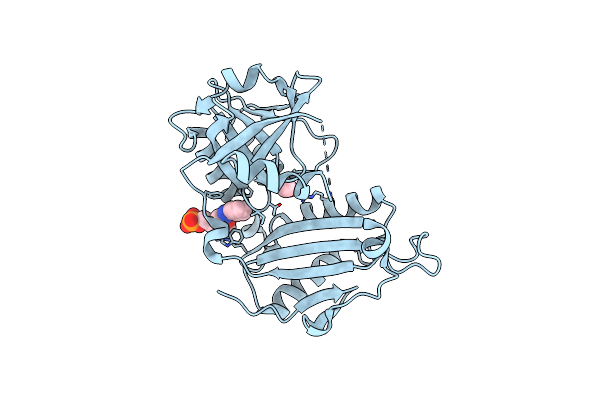 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis (Apo Form) After 15 Sec Of Soaking With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: ACT, ZXN |
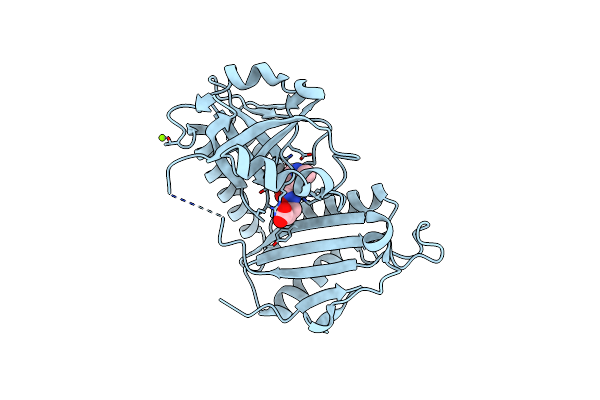 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Complexed With 3-Aminooxypropionic Acid
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: OCF, MG |
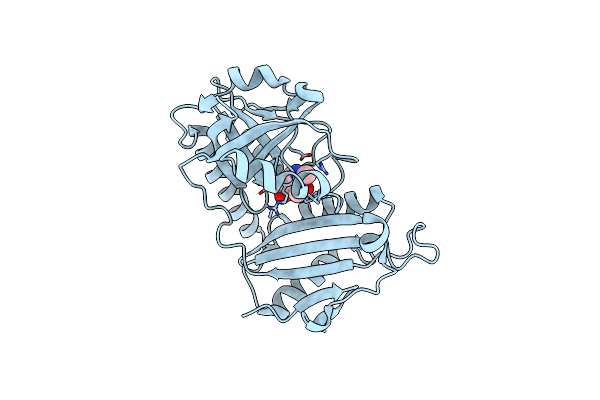 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I (Holo Form)
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I Complexed With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: ZXN, GOL |
 |
Crystal Structure Of D-Amino Acid Aminotrensferase From Haliscomenobacter Hydrossis Complexed With D-Cycloserine
Organism: Haliscomenobacter hydrossis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: GOL, LCS |
 |
Crystal Structure Of D-Aminoacid Transaminase From Haliscomenobacter Hydrossis In Its Intermediate Form
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: SO4, MG |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis (Holo Form).
Organism: Haliscomenobacter hydrossis (strain atcc 27775 / dsm 1100 / lmg 10767 / o)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-28 Classification: TRANSFERASE Ligands: PLP, PO4, ACT |
 |
Crystal Structure Of Haliscomenobacter Hydrossis Iodotyrosine Deiodinase (Iyd) Bound To Fmn
Organism: Haliscomenobacter hydrossis (strain atcc 27775 / dsm 1100 / lmg 10767 / o)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Haliscomenobacter Hydrossis Iodotyrosine Deiodinase (Iyd) Bound To Fmn And Mono-Iodotyrosine (I-Tyr)
Organism: Haliscomenobacter hydrossis (strain atcc 27775 / dsm 1100 / lmg 10767 / o)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, IYR |
 |
Crystal Structure Of Haliscomenobacter Hydrossis Iodotyrosine Deiodinase (Iyd) Bound To Fmn And 2-Iodophenol (2Ip)
Organism: Haliscomenobacter hydrossis (strain atcc 27775 / dsm 1100 / lmg 10767 / o)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, 6X8 |

