Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
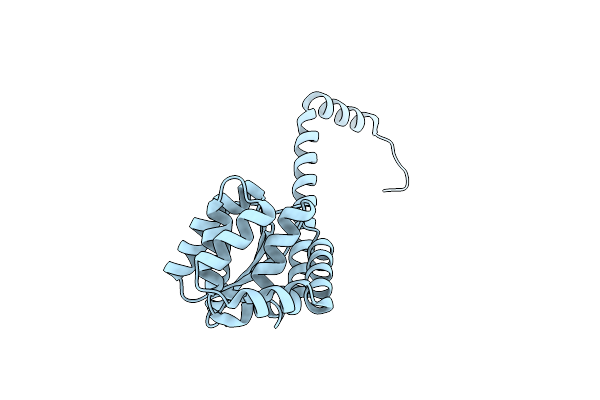 |
Structure Of Caulobacter Crescentus Suppressor Of Copper Sensitivity Protein C
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE |
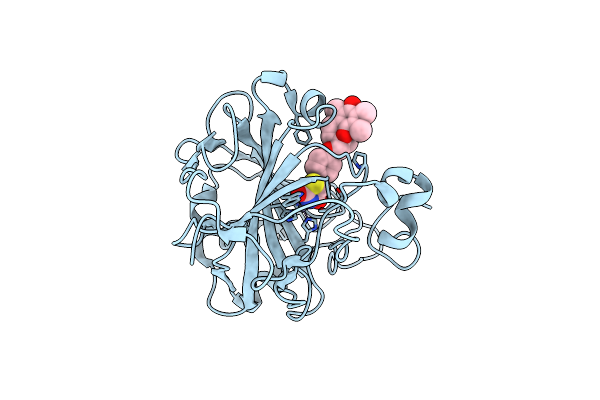 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-02-02 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, TDZ |
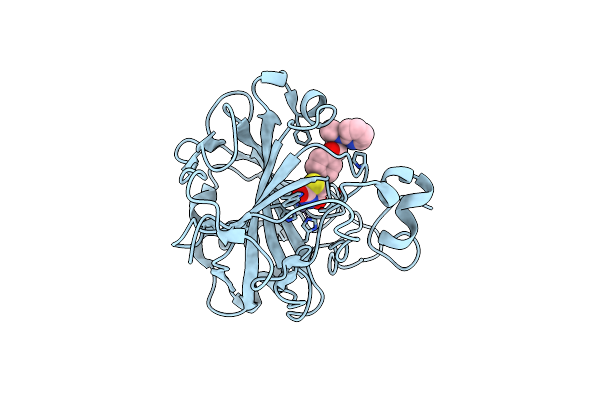 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-02-02 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, RGZ |
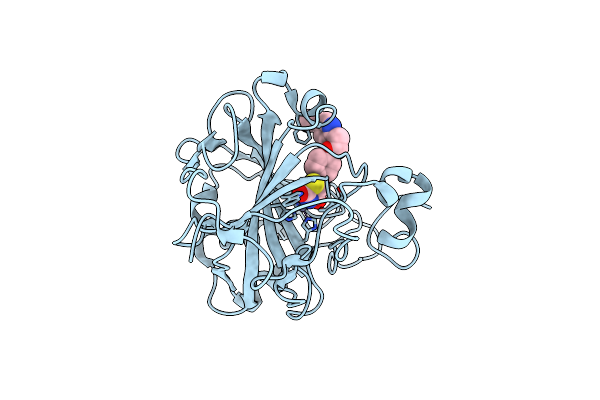 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-02-02 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, P1B |
 |
Burkholderia Pseudomallei Disulfide Bond Forming Protein A (Dsba) Liganded With Fragment Bromophenoxy Propanamide
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: YCS |
 |
Burkholderia Pseudomallei Disulfide Bond Forming Protein A (Dsba) Liganded With Fragment 4-Methoxy-N-Phenylbenzenesulfonamide
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: YCY, SO4 |
 |
Crystal Structure Of The Thiol-Disulfide Exchange Protein Alpha-Dsba2 From Wolbachia Pipientis
Organism: Wolbachia endosymbiont of drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-04-17 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2017-08-02 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2017-07-26 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-08 Classification: ISOMERASE |
 |
The Complex Structure Of E. Coli Dsba Bound To A Peptide At The Dsba/Dsbb Interface
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-01-14 Classification: OXIDOREDUCTASE/PEPTIDE INHIBITOR Ligands: ACE, NH2 |
 |
Organism: Acinetobacter baumannii aye, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-06-18 Classification: TRANSLATION/OXIDOREDUCTASE Ligands: MG, GDP, GOL |
 |
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2013-12-11 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Disulfide Oxidoreductase From Klebsiella Pneumoniae In Reduced State
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Disulfide Oxidoreductase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-10-02 Classification: OXIDOREDUCTASE Ligands: EPE, DIO |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-14 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Structural And Functional Studies Of Tcpg, The Vibrio Cholerae Dsba Disulfide-Forming Protein Required For Pilus And Cholera Toxin Production
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: DMS, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-01-31 Classification: HYDROLASE |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-01-31 Classification: HYDROLASE |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-01-31 Classification: HYDROLASE |