Search Count: 14
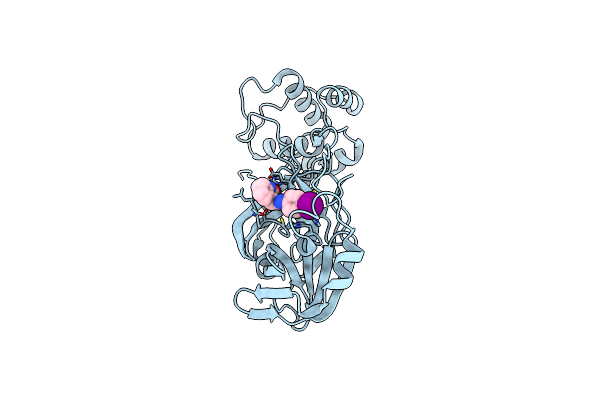 |
Identification Of Low Micromolar Sars-Cov-2 Mpro Inhibitors From Hits Identified By In Silico Screens
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: 4N0 |
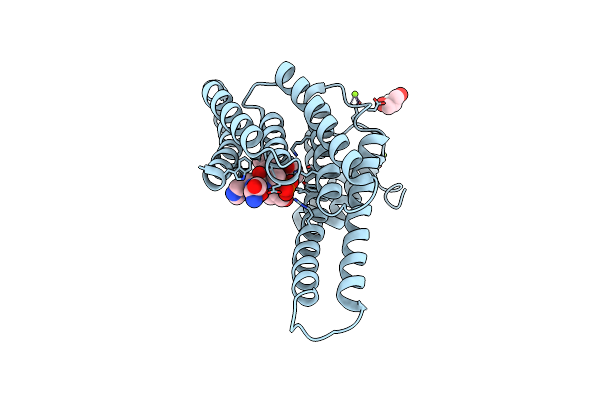 |
A Novel Phospho-Switch In The Linker Region Of The Snail Zinc Finger Protein Which Regulates 14-3-3 Association, Dna Binding And Epithelial-Mesenchymal Differentiation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-06-17 Classification: SIGNALING PROTEIN Ligands: MG, GOL |
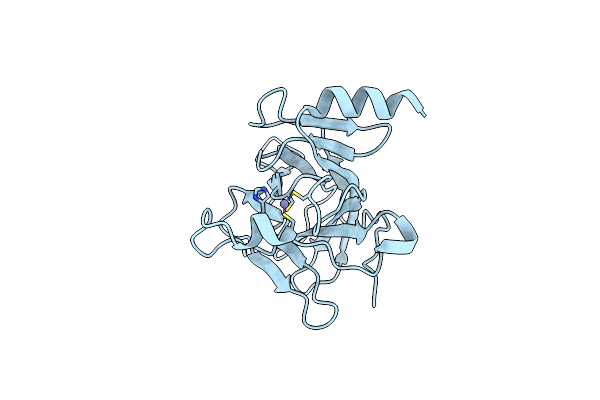 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-01-15 Classification: ANTITUMOR PROTEIN Ligands: ZN |
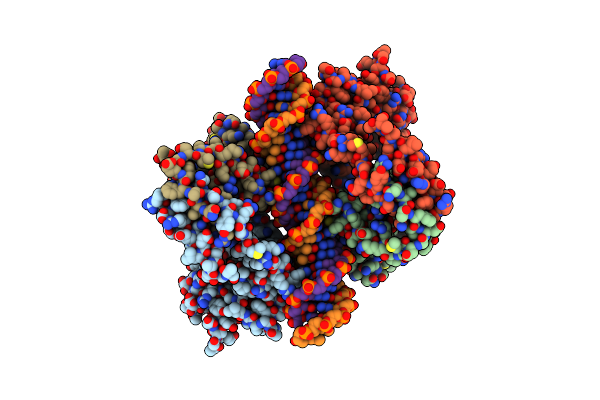 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-01-15 Classification: TRANSCRIPTION/DNA Ligands: ZN |
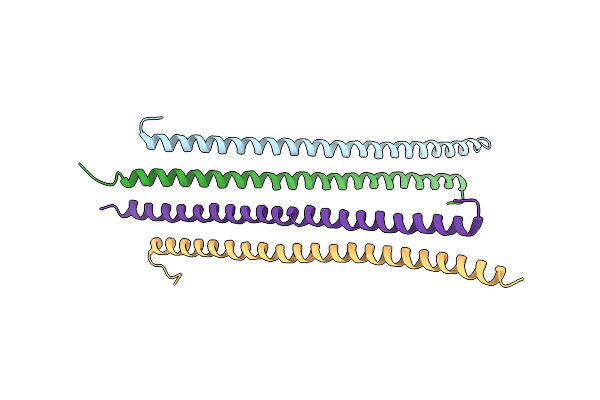 |
Structural Basis Of Transcriptional Gene Silencing Mediated By Arabidopsis Mom1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-03-07 Classification: TRANSCRIPTION |
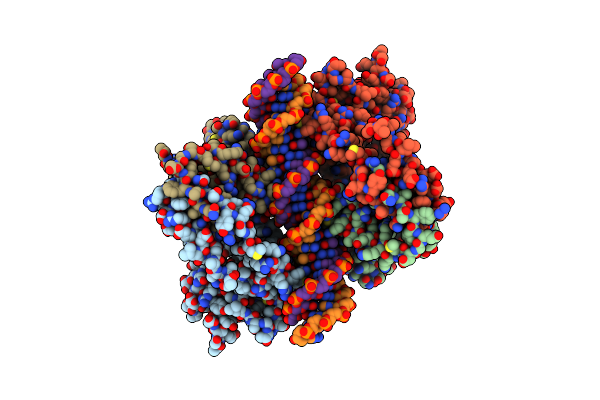 |
Crystal Structure Of A Multidomain Human P53 Tetramer Bound To The Natural Cdkn1A(P21) P53-Response Element
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-11-23 Classification: ANTITUMOR PROTEIN/DNA Ligands: ZN |
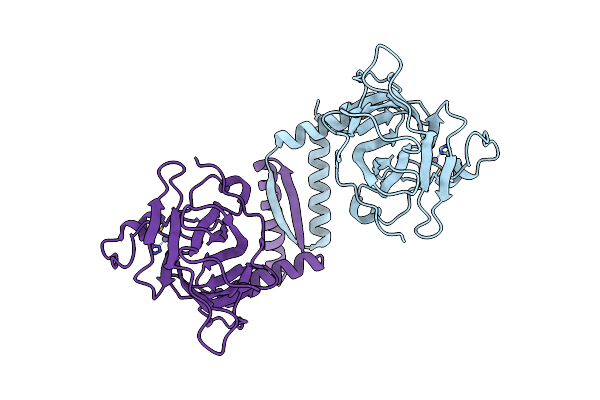 |
An Induced Fit Mechanism Regulates P53 Dna Binding Kinetics To Confer Sequence Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-11 Classification: ANTITUMOR PROTEIN Ligands: ZN |
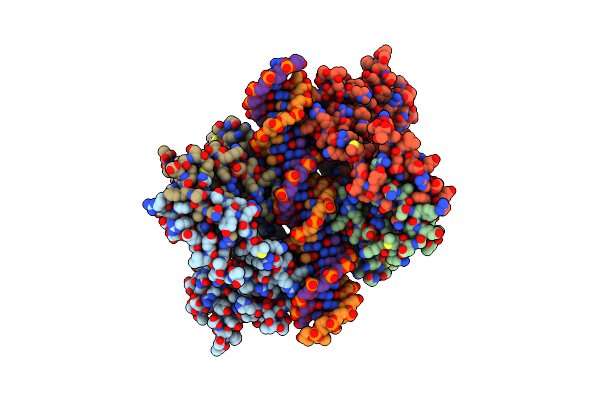 |
An Induced Fit Mechanism Regulates P53 Dna Binding Kinetics To Confer Sequence Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-05-11 Classification: ANTITUMOR PROTEIN/DNA Ligands: ZN |
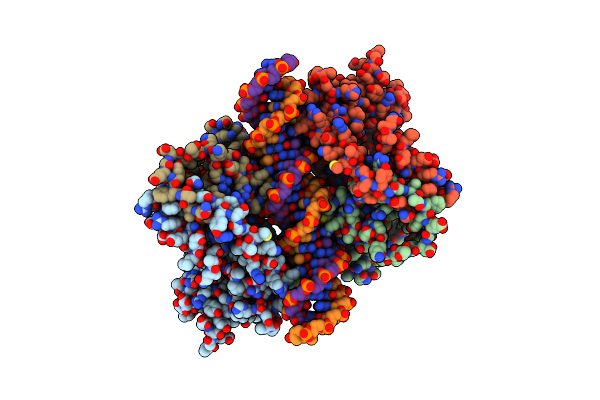 |
An Induced Fit Mechanism Regulates P53 Dna Binding Kinetics To Confer Sequence Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2011-05-11 Classification: CELL CYCLE/DNA Ligands: ZN |
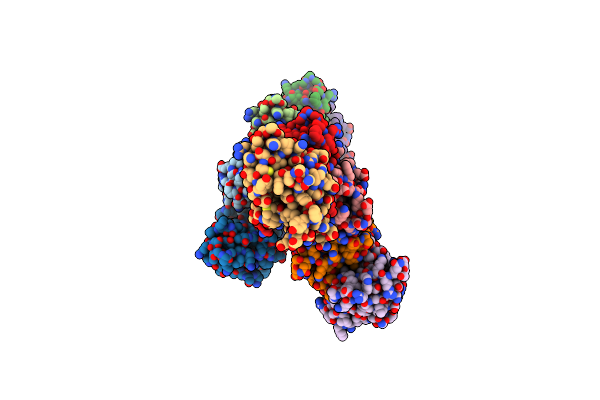 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-11-30 Classification: CELL CYCLE |
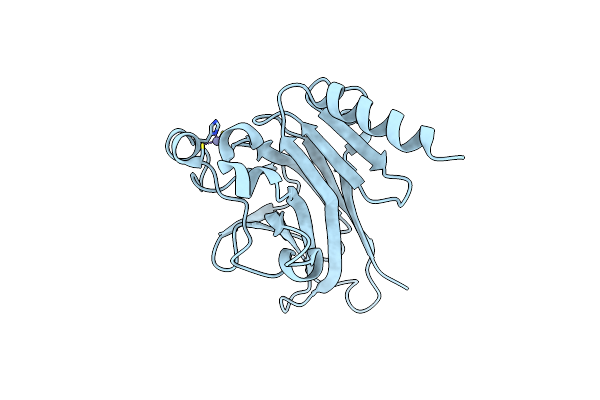 |
Structural Differences In The Dna Binding Domains Of Human P53 And Its C. Elegans Ortholog Cep-1: Structure Of C. Elegans Cep-1
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-07-20 Classification: TRANSCRIPTION Ligands: ZN |
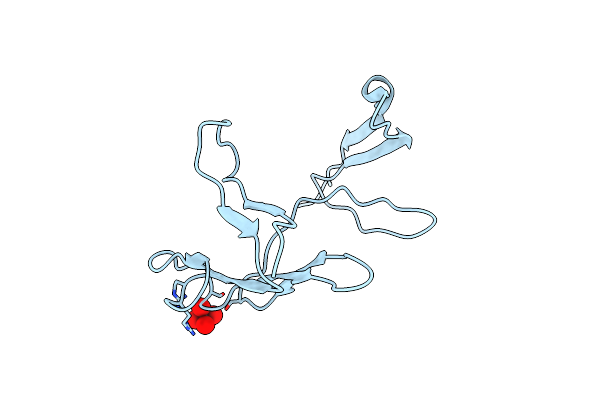 |
Crystal Structure Of The Fha Domain Of The Chfr Mitotic Checkpoint Protein Complexed With Tungstate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-05-08 Classification: CELL CYCLE Ligands: WO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-05-08 Classification: CELL CYCLE |
 |
Solution Structure Determination Of A P53 Mutant Dimerization Domain, Nmr, Minimized Average Structure
|

