Search Count: 50
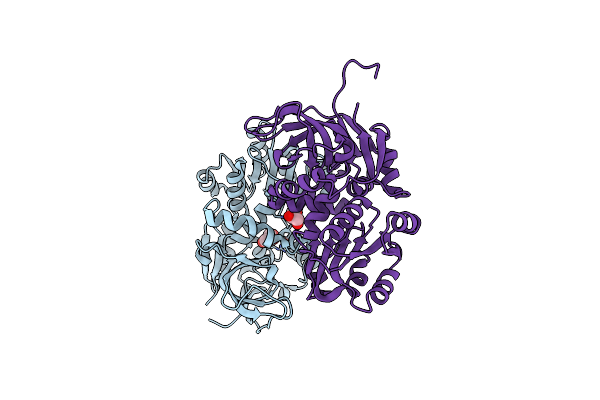 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-11-18 Classification: OXIDOREDUCTASE Ligands: GOL |
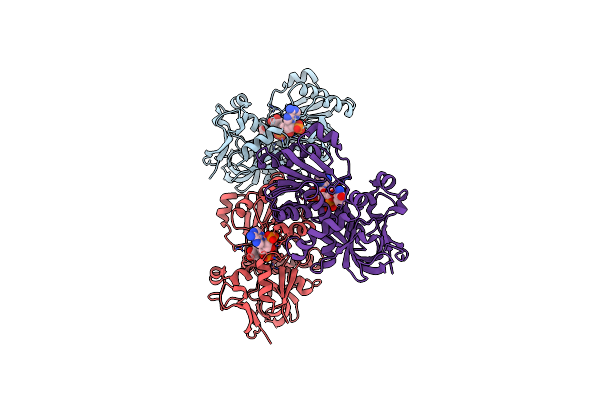 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2015-11-18 Classification: OXIDOREDUCTASE Ligands: ACT, NDP |
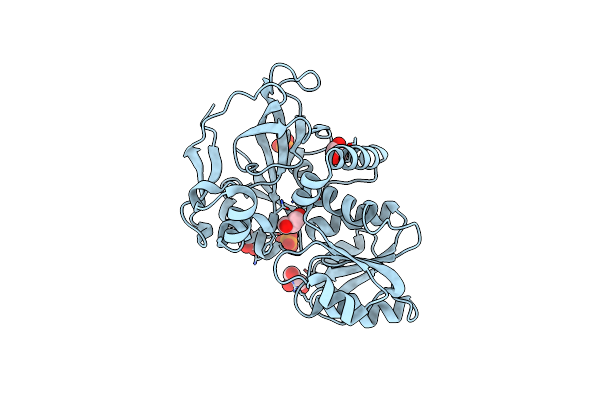 |
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-11-18 Classification: OXIDOREDUCTASE Ligands: GOL, PO4 |
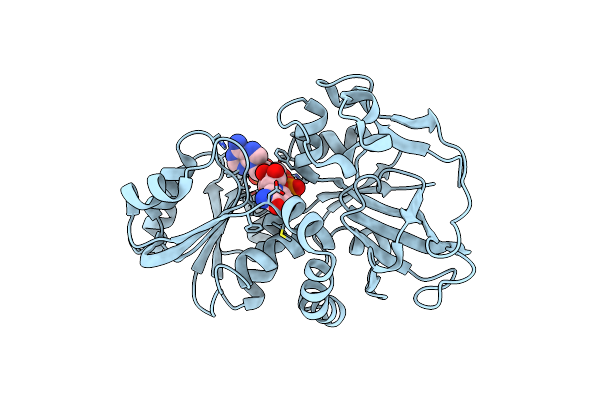 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2015-11-18 Classification: OXIDOREDUCTASE Ligands: NAP |
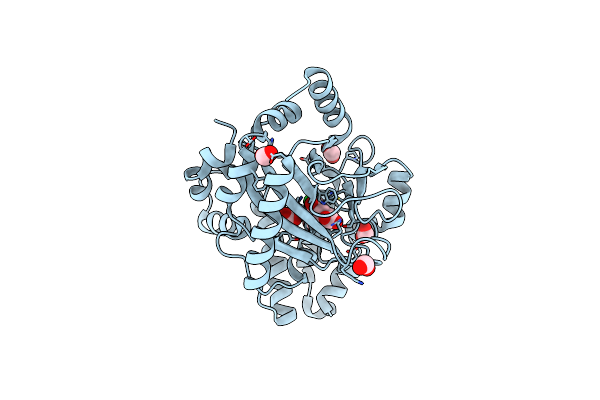 |
Halogenase Domain From Cura Module With Fe, Chloride, And Alpha-Ketoglutarate
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-07-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, FMT, AKG, CL |
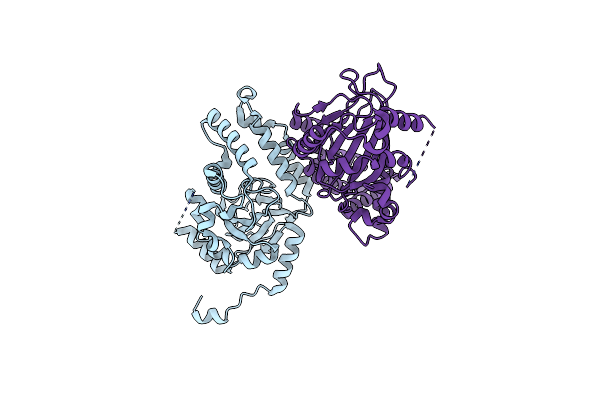 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-07-28 Classification: BIOSYNTHETIC PROTEIN |
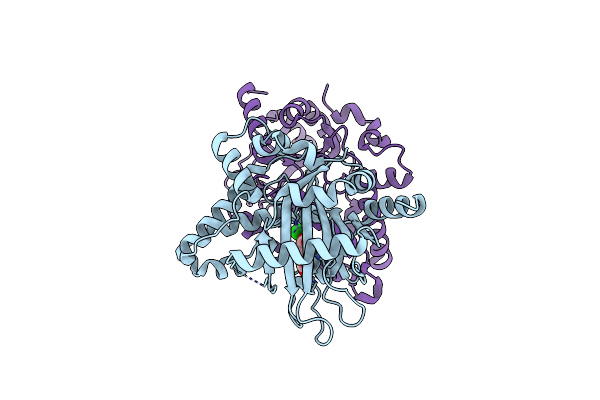 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2010-07-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, AKG, CL |
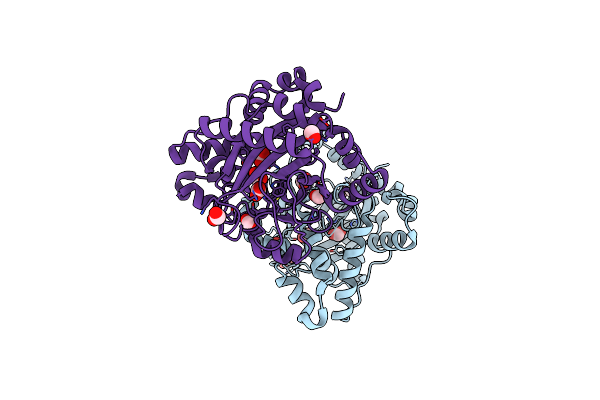 |
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2010-07-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-12-15 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Loading Gnatl Domain Of Cura From Lyngbya Majuscula
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-11-20 Classification: TRANSFERASE, LYASE Ligands: SO4, CL, GOL |
 |
Crystal Structure Of The Loading Gnatl Domain Of Cura From Lyngbya Majuscula Soaked With Malonyl-Coa
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2007-11-20 Classification: TRANSFERASE, LYASE Ligands: ACO |
 |
Crystal Structure Of The Ech2 Decarboxylase Domain Of Curf From Lyngbya Majuscula
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-09 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of The Ech2 Decarboxylase Domain Of Curf From Lyngbya Majuscula, Rhombohedral Crystal Form
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-10-09 Classification: LYASE |
 |
Crystal Structure Of The Y82F Variant Of Ech2 Decarboxylase Domain Of Curf From Lyngbya Majuscula
Organism: Lyngbya majuscula
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-10-09 Classification: LYASE Ligands: CL, GOL |
 |
Organism: Aequorea victoria, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-12-20 Classification: ELECTRON TRANSPORT Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2006-12-11 Classification: ELECTRON TRANSPORT Ligands: GSH |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: NI |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-05-04 Classification: OXIDOREDUCTASE Ligands: SO4, CL, HEM |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-03-13 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2001-01-10 Classification: HYDROLASE Ligands: CD |

