Search Count: 38
 |
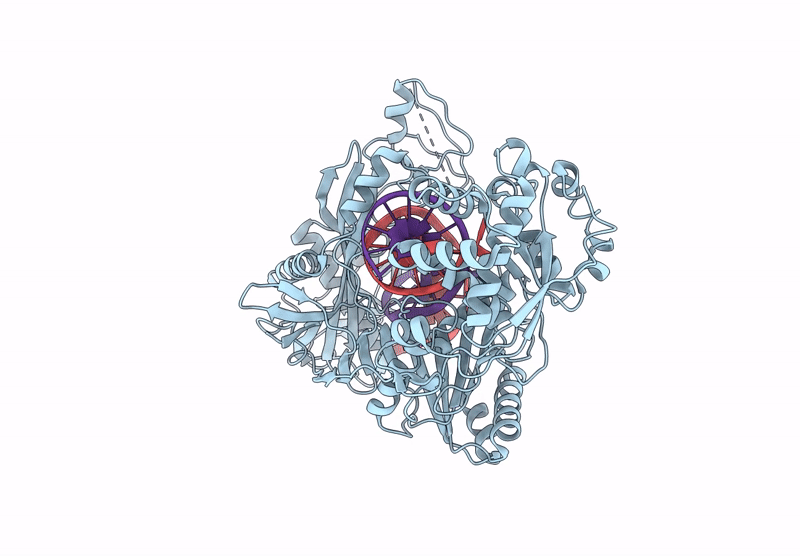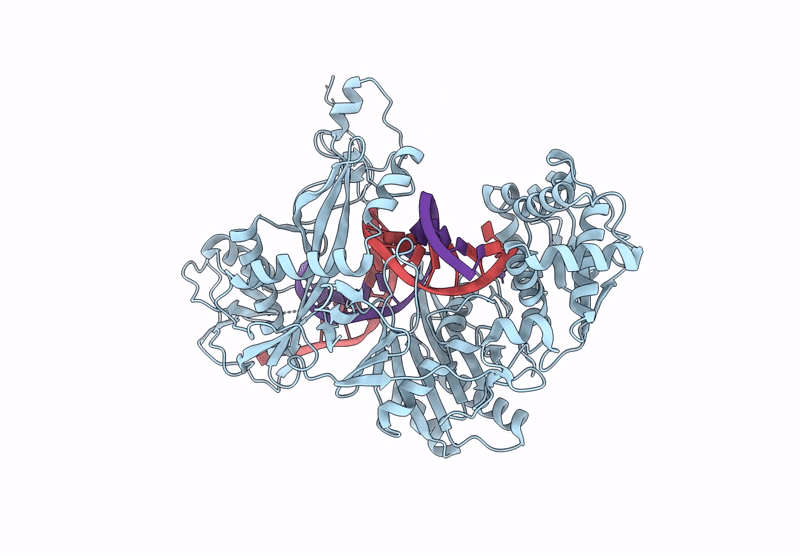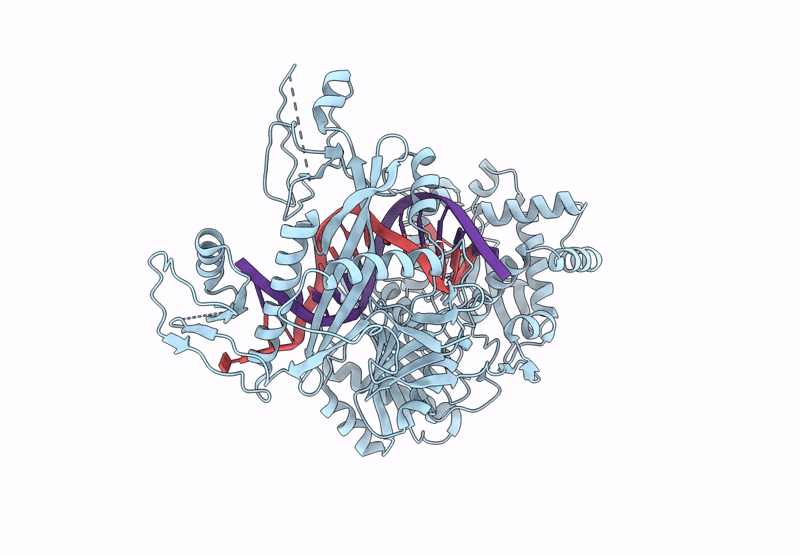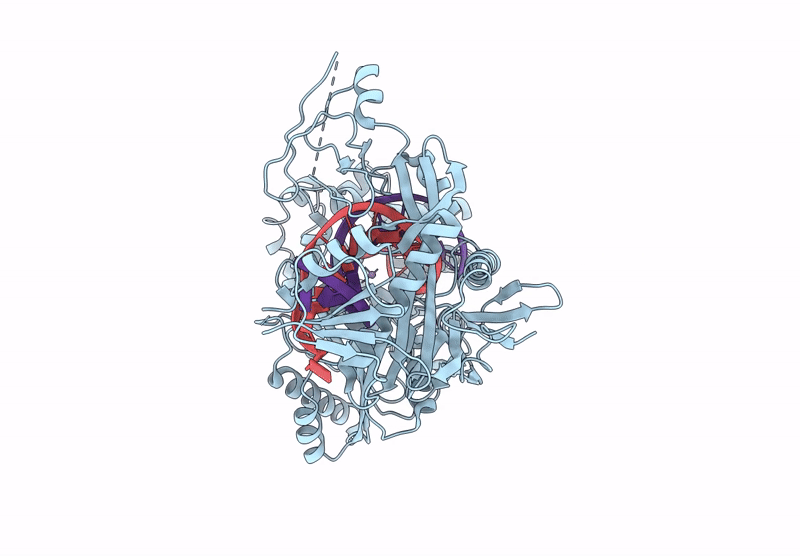
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
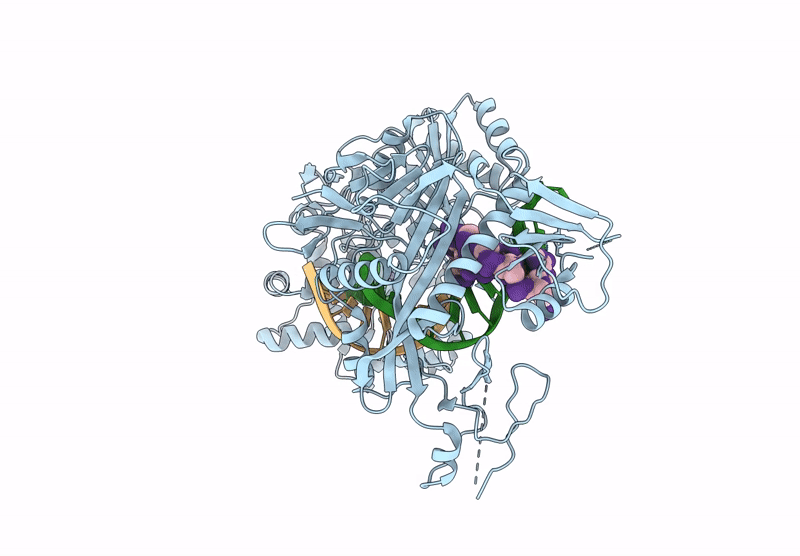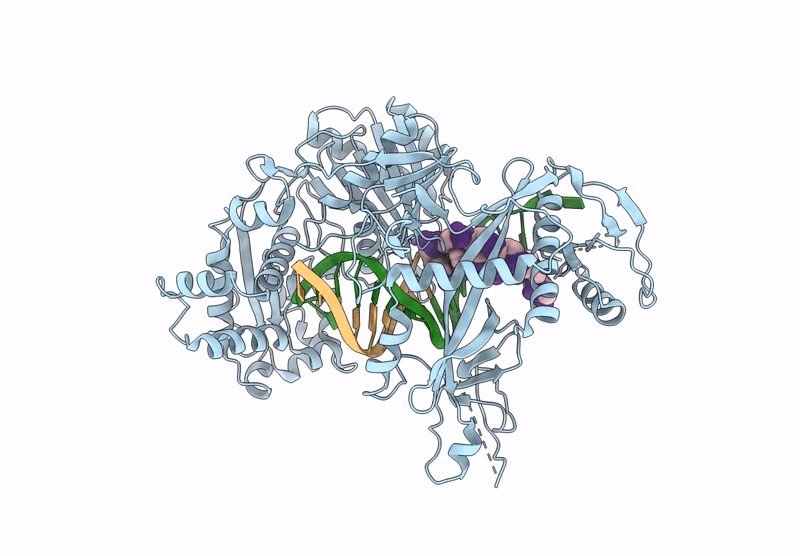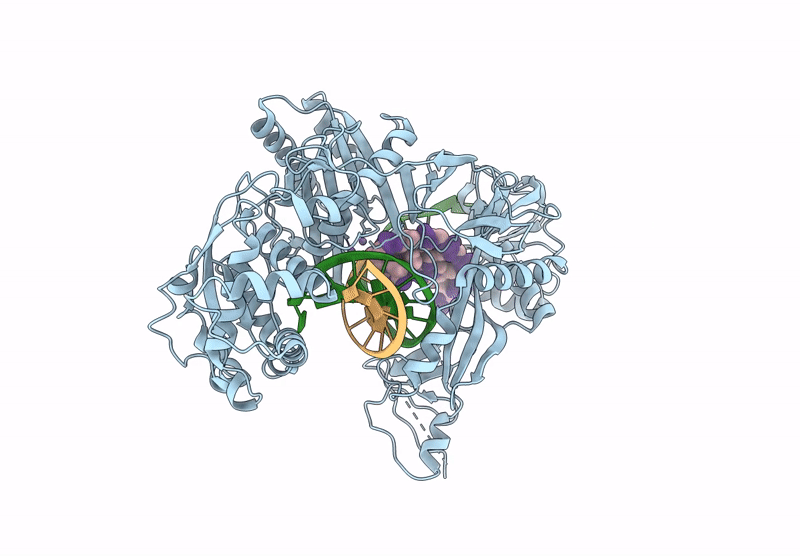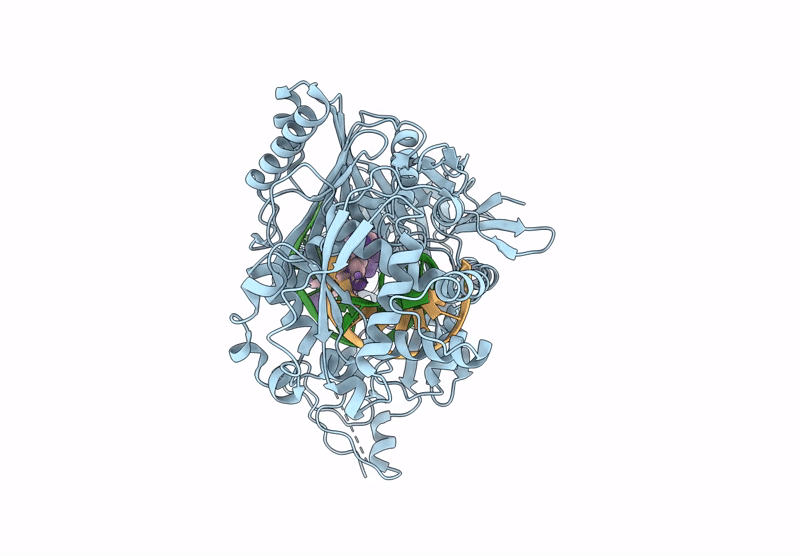
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
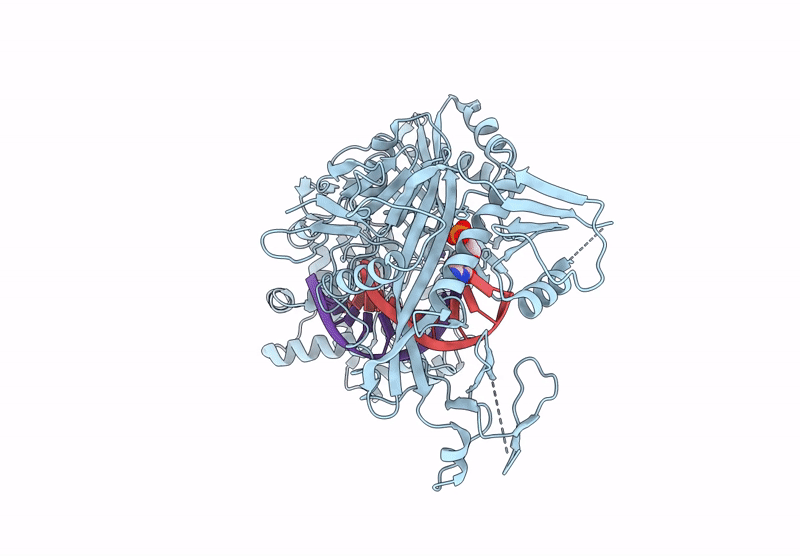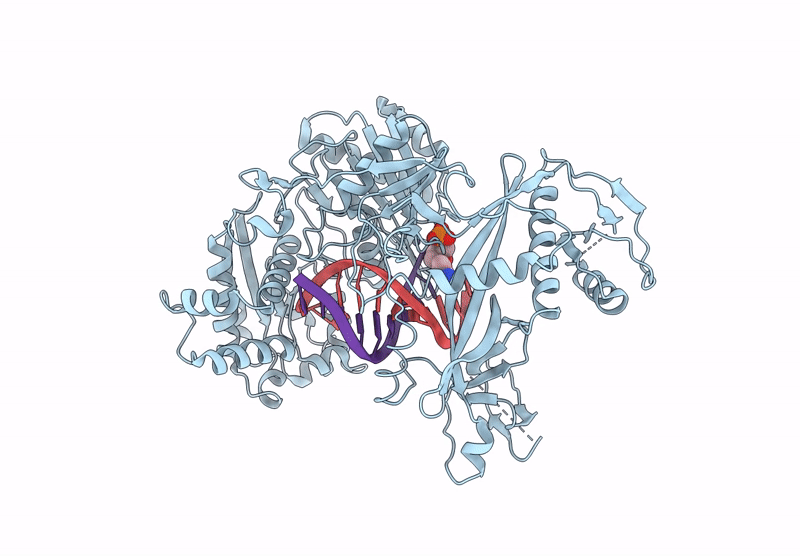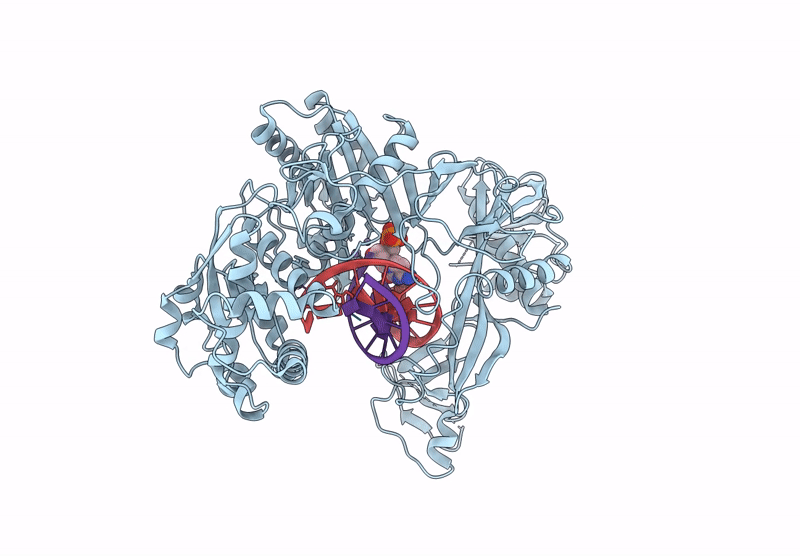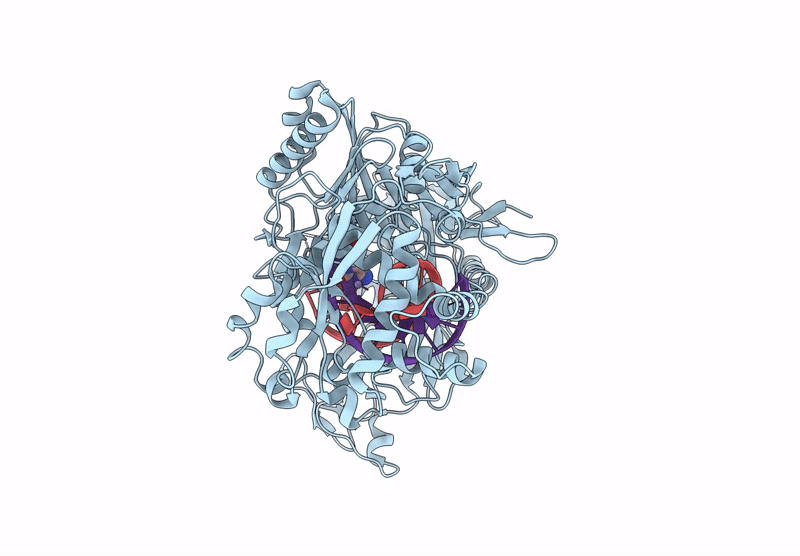
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Target-Cleaved State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Target-Released State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, DC |
 |
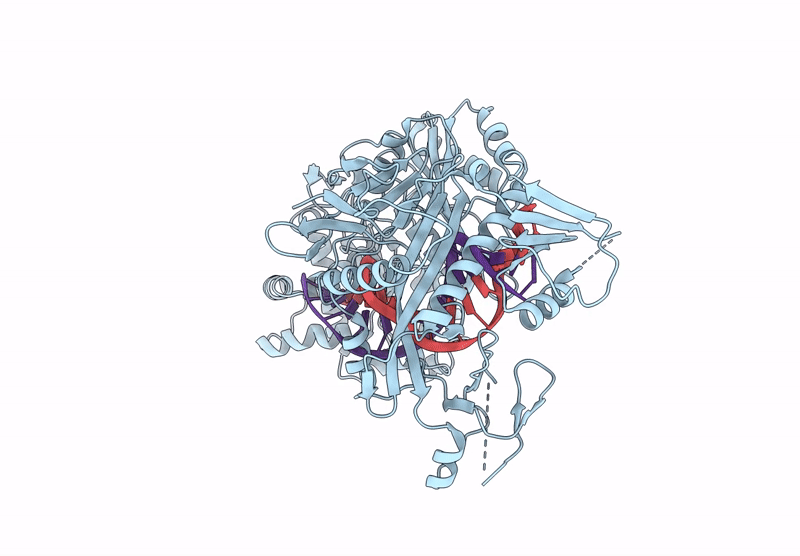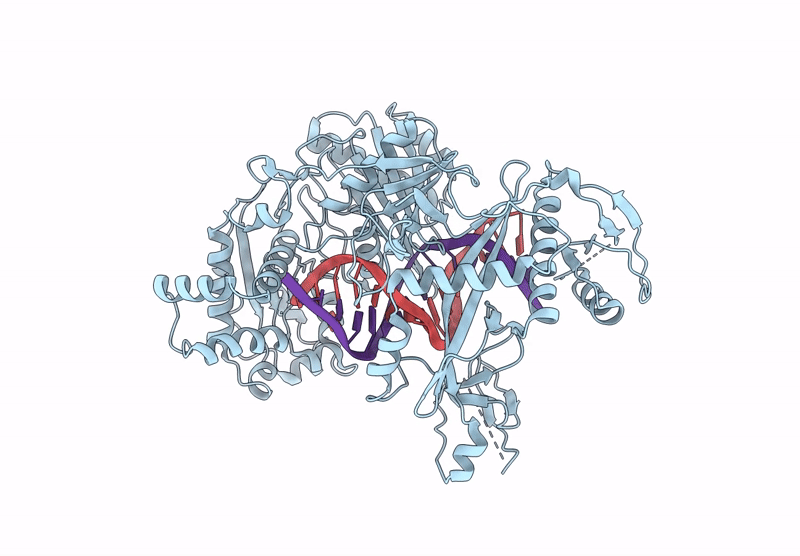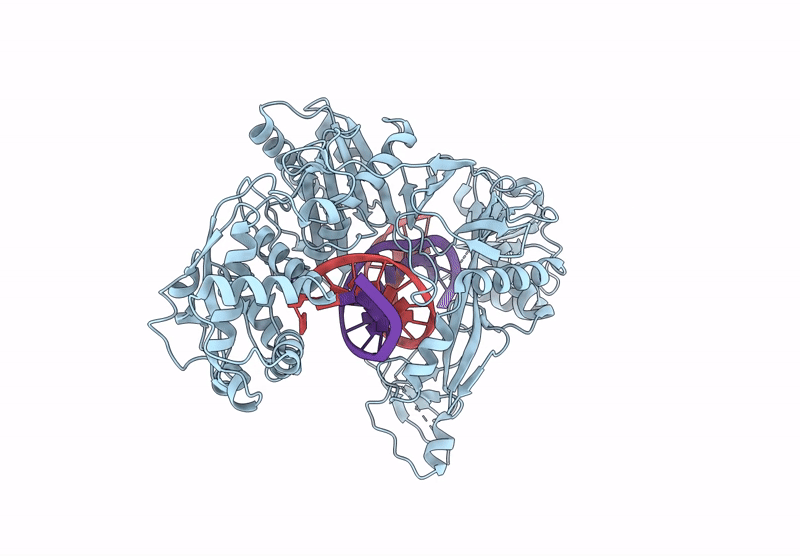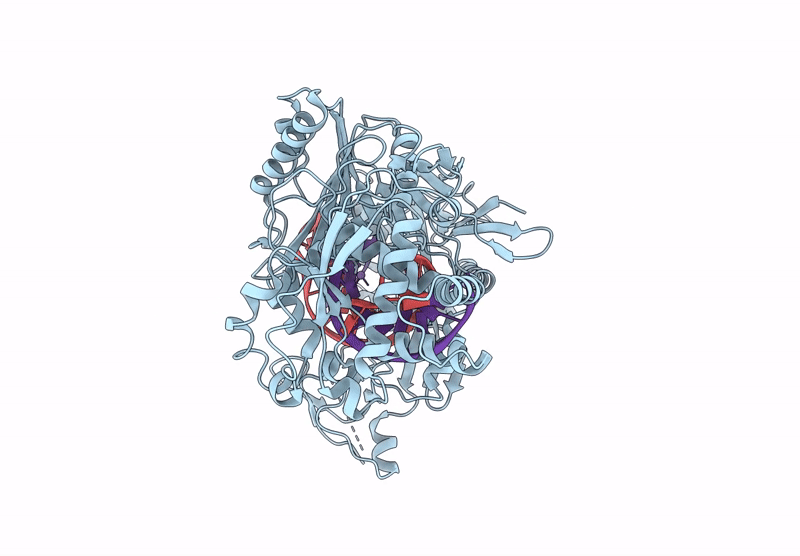
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: DNA BINDING PROTEIN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Rna
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Structure Of The Argonaute Protein From Kurthia Massiliensis In Complex With Guide Dna And 19-Mer Target Dna In Pre-Cleavage State
Organism: Kurthia massiliensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: DD9, D10, C14, HP6, D12, XKP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: DD9, D10, C14, HP6, D12, XKP |
 |
Organism: Mus musculus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-08-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, G70, ACP |
 |
Organism: Mus musculus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-08-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, CL, GDP, MES, GDF |
 |
Organism: Mus musculus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-08-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, CL, GDP, MES, HOS, ACP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-19 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Apo Form Fibronectin-Binding Protein Apa From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-29 Classification: PROTEIN BINDING Ligands: GOL |
 |
Crystal Structure Of Fibronectin-Binding Protein Apa Mutant From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2019-05-29 Classification: PROTEIN BINDING Ligands: HG, GOL |
 |
Cdk8-Cycc In Complex With 8-[3-(3-Amino-1H-Indazol-6-Yl)-5-Chloro- Pyridine-4-Yl]-2,8-Diaza-Spiro[4.5]Decan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: 5XG, DMS, EDO, FMT |
 |
Cdk8-Cycc In Complex With 8-[3-Chloro-5-(1-Methyl-2,2-Dioxo-2, 3-Dihydro-1H-2L6-Benzo[C]Isothiazol-5-Yl)-Pyridin- 4-Yl]-1-Oxa-3,8-Diaza-Spiro[4.5]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: 5Y6, DMS, EDO, FMT |
 |
Cdk8-Cycc In Complex With 5-{5-Chloro-4-[1-(2-Methoxy-Ethyl)-1,8-Diaza-Spiro[4.5]Dec-8-Yl]-Pyridin-3-Yl}-1-Methyl-1,3-Dihydro-Benzo[C]Isothiazole 2,2-Dioxide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: 5Y7, EDO, FMT |
 |
Cdk8-Cycc In Complex With 8-[2-Amino-3-Chloro-5-(1-Methyl-1H-Indazol-5-Yl)-Pyridin-4-Yl]-2,8-Diaza-Spiro[4.5]Decan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: 5Y8, EDO, FMT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: ZN, 5TZ, EDO, DMS, CL |

