Search Count: 20
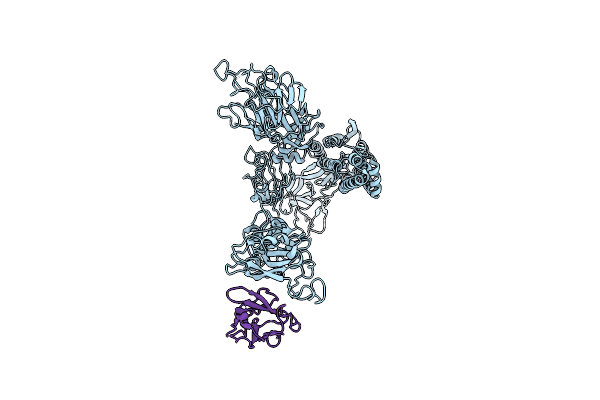 |
Minor Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874A Vhh, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
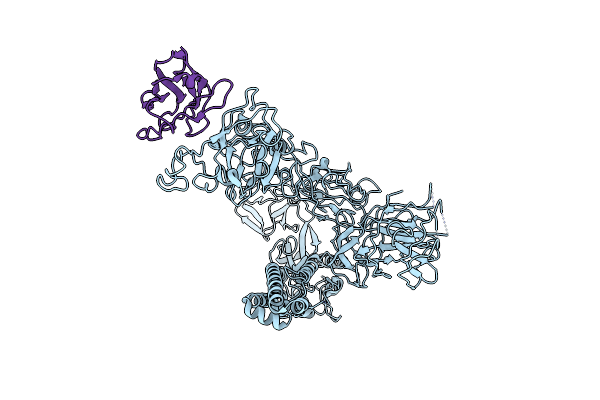 |
Major Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
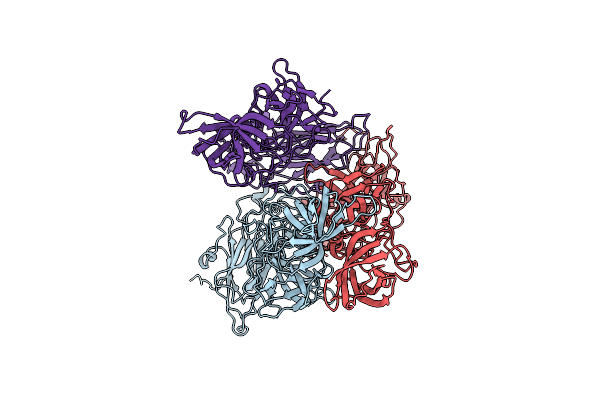
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
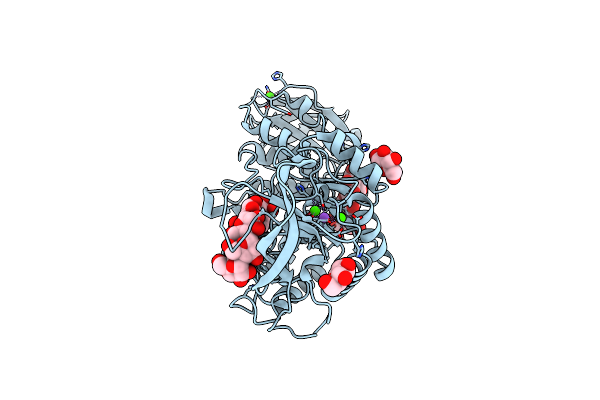
 |
Organism: Murine norovirus gv/nih-2410/2005/usa
Method: ELECTRON MICROSCOPY Release Date: 2020-02-26 Classification: VIRUS |
 |
Structure Of The Human M2 Muscarinic Acetylcholine Receptor Bound To An Antagonist
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-02-01 Classification: SIGNALING PROTEIN/ANTAGONIST Ligands: QNB, BGC, CL |
 |
Crystal Structure Of Maltohexaose-Producing Amylase From Bacillus Sp.707 Complexed With Maltopentaose.
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: GLC, CA, NA |
 |
Crystal Structure Of Maltohexaose-Producing Amylase From Bacillus Sp.707 Complexed With Maltohexaose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: GLC, CA, NA |
 |
Crystal Structure Of Maltohexaose-Producing Amylase From Alkalophilic Bacillus Sp.707.
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-11-30 Classification: HYDROLASE Ligands: CA, NA, PO4, TRS |
 |
Crystal Structure Of Maltohexaose-Producing Amylase Complexed With Pseudo-Maltononaose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-11-30 Classification: HYDROLASE Ligands: ACI, CA, NA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-08-03 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-08-03 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of F283L Mutant Cyclodextrin Glycosyltransferase Complexed With A Pseudo-Tetraose Derived From Acarbose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-08-03 Classification: TRANSFERASE Ligands: GLC, ACI, CA |
 |
Crystal Structure Of F283Y Mutant Cyclodextrin Glycosyltransferase Complexed With A Pseudo-Tetraose Derived From Acarbose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-08-03 Classification: TRANSFERASE Ligands: GLC, ACI, GAL, CA |
 |
Crystal Structure Of Cyclodextrin Glucanotransferase Complexed With A Pseudo-Maltotetraose Derived From Acarbose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-02-24 Classification: TRANSFERASE Ligands: GLC, ACI, CA |
 |
Crystal Structure Of F183L/F259L Mutant Cyclodextrin Glucanotransferase Complexed With A Pseudo-Maltotetraose Derived From Acarbose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-02-24 Classification: TRANSFERASE Ligands: GLC, ACI, CA |
 |
Crystal Structure Of Y100L Mutant Cyclodextrin Glucanotransferase Compexed With An Acarbose
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-02-24 Classification: TRANSFERASE Ligands: ACI, CA |
 |
Crystal Structure Of Cyclodextrin Glucanotransferase From Alkalophilic Bacillus Sp.#1011 Complexed With 1-Deoxynojirimycin
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-04-11 Classification: TRANSFERASE Ligands: CA, NOJ |
 |
Crystal Structure Of Alkalophilic Asparagine 233-Replaced Cyclodextrin Glucanotransferase Complexed With An Inhibitor, Acarbose, At 2.0 A Resolution
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-04-07 Classification: TRANSFERASE Ligands: CA |
 |
Crystal Structure Of Asparagine 233-Replaced Cyclodextrin Glucanotransferase From Alkalophilic Bacillus Sp. 1011 Determined At 1.9 A Resolution
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-03-17 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-01-11 Classification: GLYCOSYLTRANSFERASE Ligands: CA |

