Search Count: 24
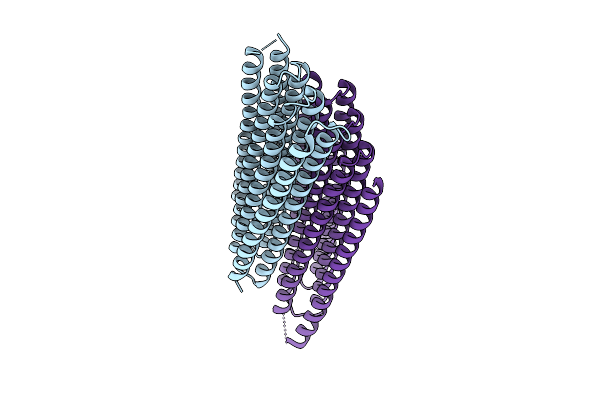 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN |
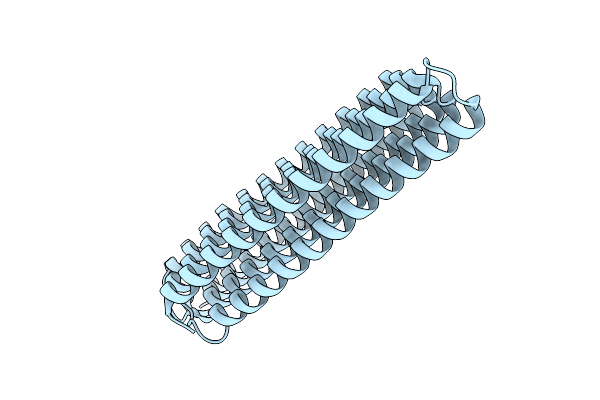 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN |
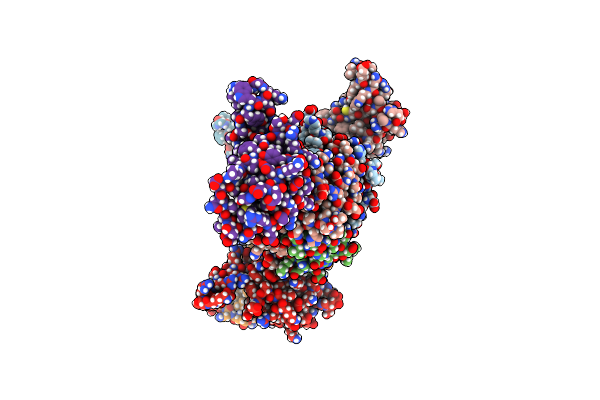 |
Crystal Structure Of T33-18.2: Deep-Learning Sequence Design Of Co-Assembling Tetrahedron Protein Nanoparticles
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN |
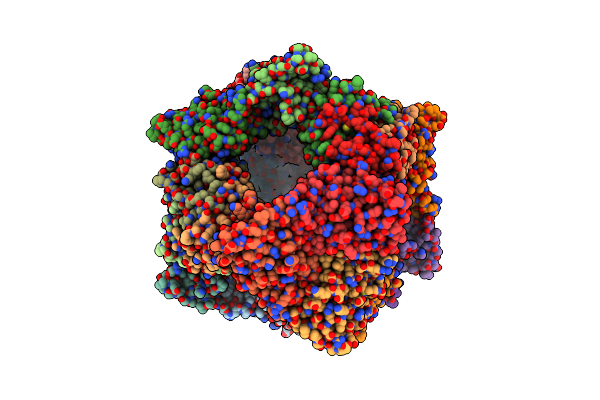 |
Crystal Structure Of T33-28.3: Deep-Learning Sequence Design Of Co-Assembling Tetrahedron Protein Nanoparticles
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of T33-27.1: Deep-Learning Sequence Design Of Co-Assembling Tetrahedron Protein Nanoparticles
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.63 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-07-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-09-28 Classification: DE NOVO PROTEIN |
 |
Organism: Helicobacter pylori, Homo sapiens, Helicobacter pylori p12
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: GOL, CL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: CELL ADHESION Ligands: GOL, CL, SO4, PEG |
 |
Organism: Homo sapiens, Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2018-05-16 Classification: CELL ADHESION |
 |
Organism: Helicobacter pylori, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2018-05-16 Classification: CELL ADHESION |
 |
Organism: Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-10-12 Classification: CELL ADHESION |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: CELL ADHESION |
 |
Crystal Structure Of The N-Terminal Domain Of Helicobacter Pylori Caga Protein
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2012-08-22 Classification: TOXIN |
 |
Crystal Structure Of Cag Virb11 (Hp0525) And An Inhibitory Protein (Hp1451)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-13 Classification: hydrolase/protein binding |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-09-25 Classification: SIGNALING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-09-25 Classification: SIGNALING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-09-25 Classification: SIGNALING PROTEIN Ligands: ACT |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, AZI |

