Search Count: 115
 |
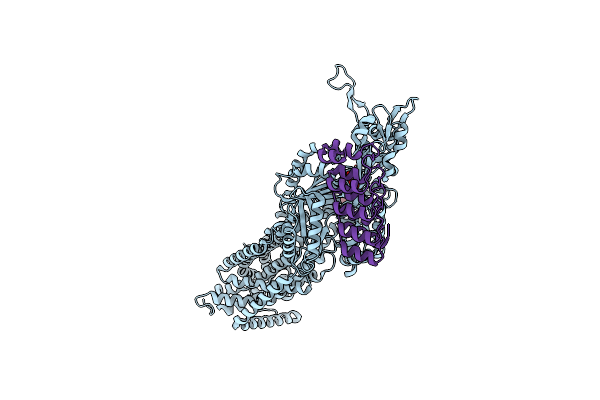
Structure Of Cx43/Gja1 Gap Junction Intercellular Channel In Complex With Dic8-Pip2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN Ligands: PIO |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
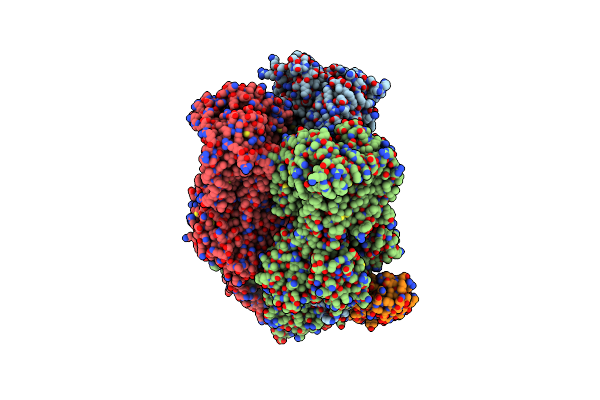 |
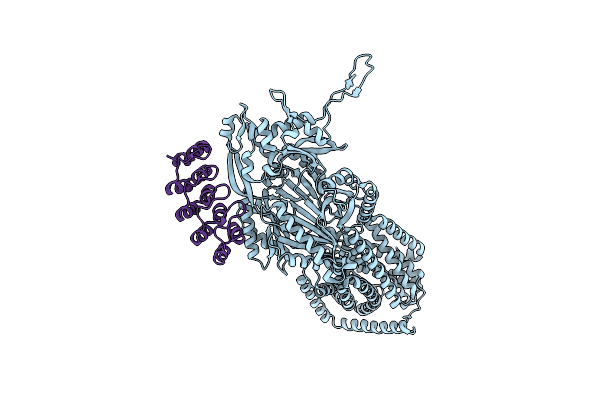
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
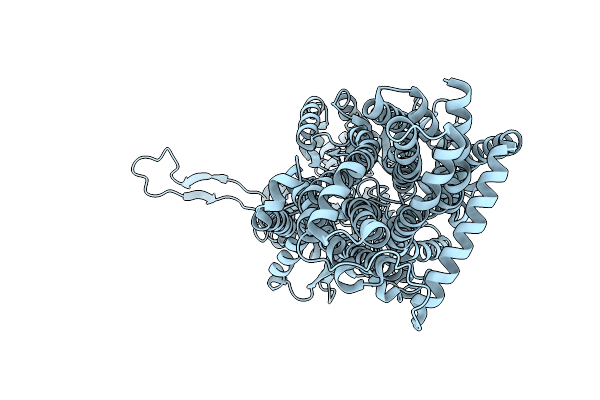
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
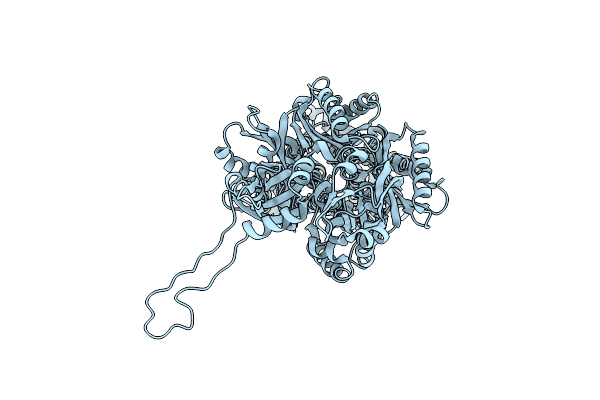
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
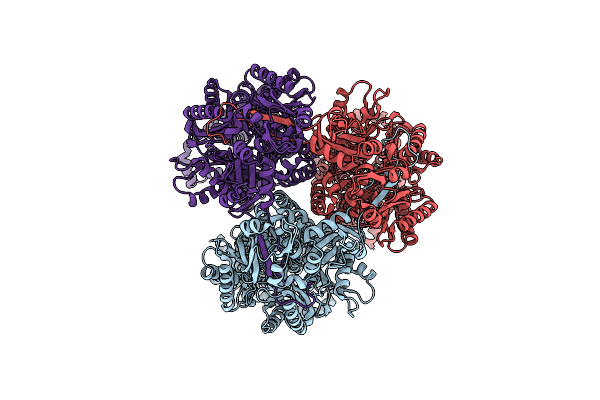
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
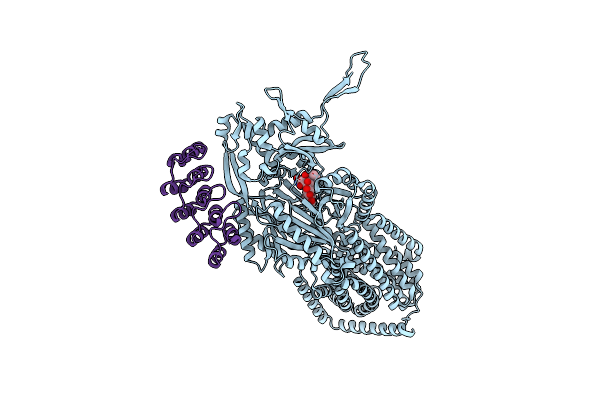
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
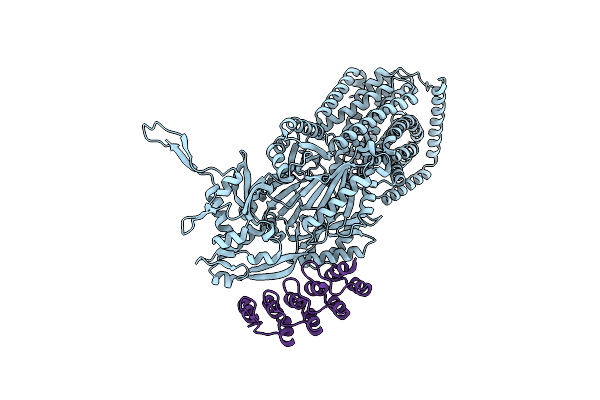
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
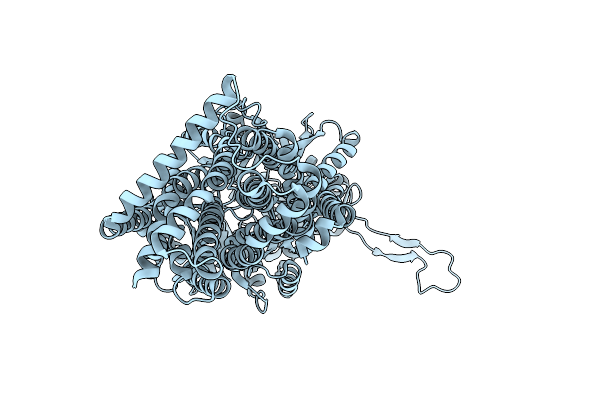 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
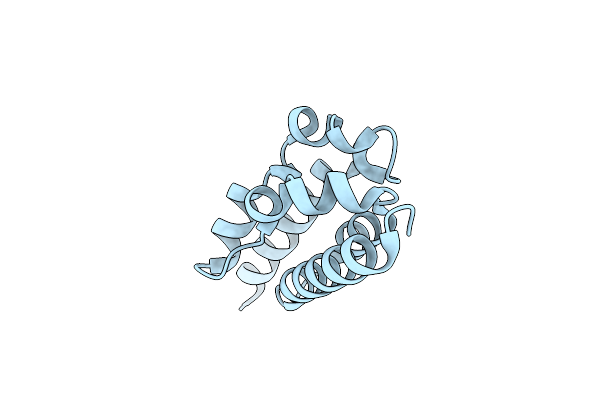 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-04-16 Classification: IMMUNE SYSTEM |
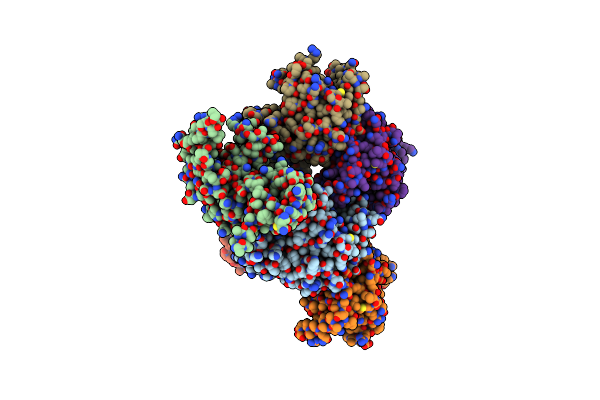 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: LYASE |
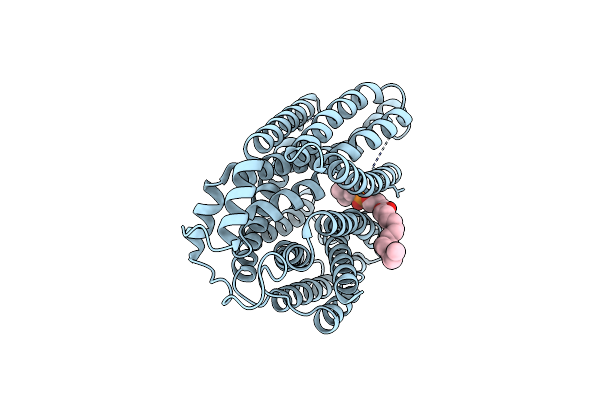 |
Organism: Aequorea victoria, Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: 42H |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-588
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BHY, ZN, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-600
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BHX, ZN, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-584
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: ZN, A1BHW, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-589
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BH8, ZN, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-599
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BH9, ZN, K |
 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
 |
Organism: Neisseria lactamica atcc 23970
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: FE |

