Search Count: 14
 |
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, PEG, GOL, BTB |
 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenensis In Complex With Fructose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, BTB, FRU, PEG |
 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenesis In Complex With Sucrose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG |
 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenensis In Complex With Raffinose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, PEG |
 |
Structure Of D169A/E171A Double Mutant Of Chitinase Chit42 From Trichoderma Harzianum Complexed With Chitinhexaose.
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ACT, ZN |
 |
Structure Of D169A/E171A Double Mutant Of Chitinase Chit42 From Trichoderma Harzianum Complexed With Chitintetraose.
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ACT, PEG, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-04-18 Classification: IMMUNE SYSTEM Ligands: 8EU |
 |
Structure Of Karilysin Mmp-Like Catalytic Domain In Complex With Inhibitory Tetrapeptide Swfp
Organism: Tannerella forsythia, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: ZN, K, NA, GOL |
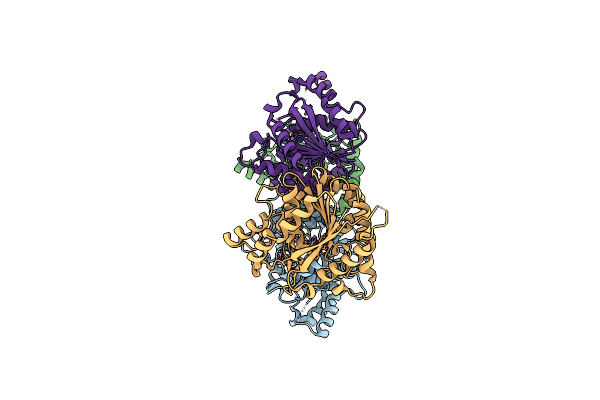 |
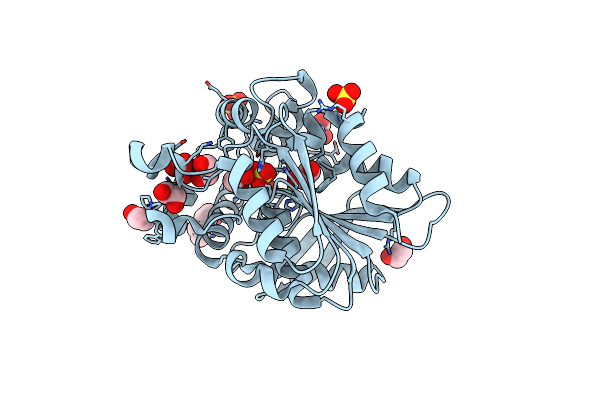
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-08-01 Classification: HYDROLASE Ligands: MN |
 |
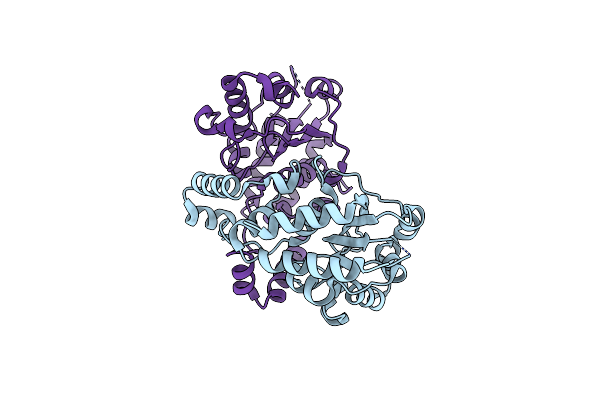
Crystal Structure Of A Probable Hydrolytic Enzyme (Pa3053) From Pseudomonas Aeruginosa Pao1 At 1.50 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: 2PE, SO4, EDO, CL |
 |
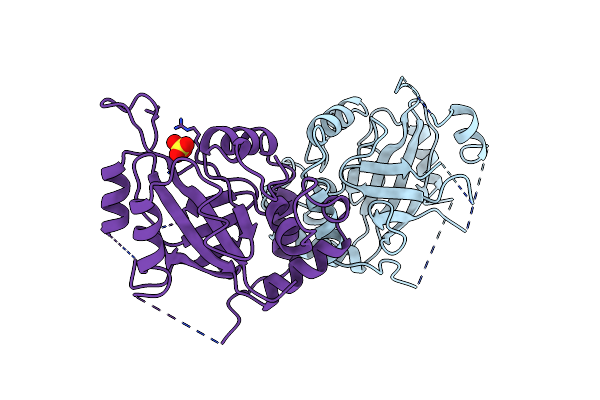
The 2.6 Angstrom Crystal Structure Of Chbp, The Cif Homologue From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2009-02-03 Classification: UNKNOWN FUNCTION |
 |
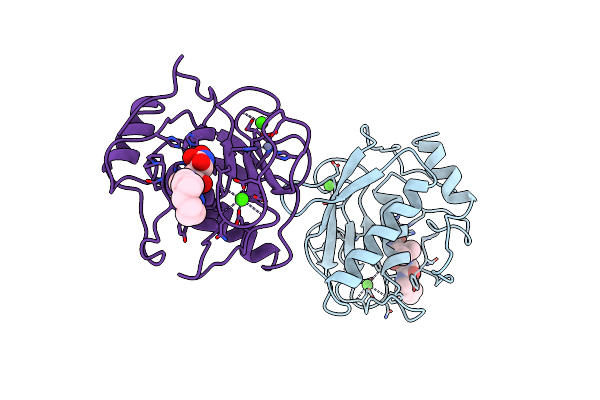
Organism: Human herpesvirus 5
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-10-15 Classification: Viral protein, hydrolase Ligands: SO4 |
 |
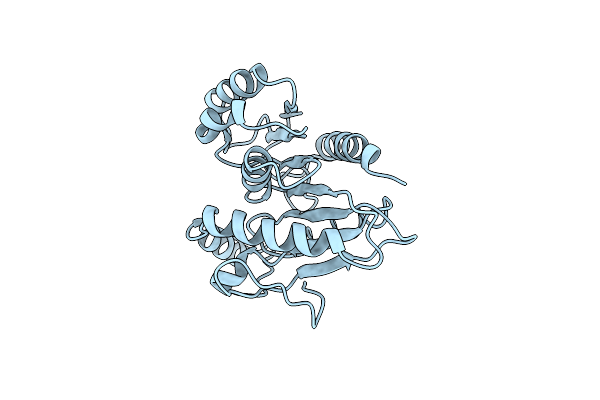
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-08-12 Classification: METALLOPROTEINASE Ligands: CA, ZN, HLE, RIN |
 |
Organism: Pseudomonas knackmussii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-08-17 Classification: HYDROLYTIC ENZYME |

