Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
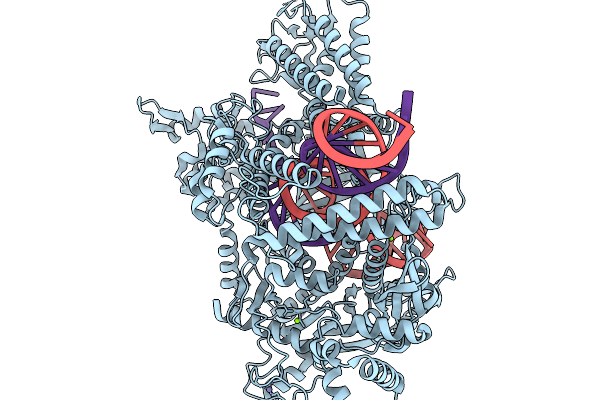
Organism: Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG |
 |
Organism: Bacteroidales, Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG, ZN |
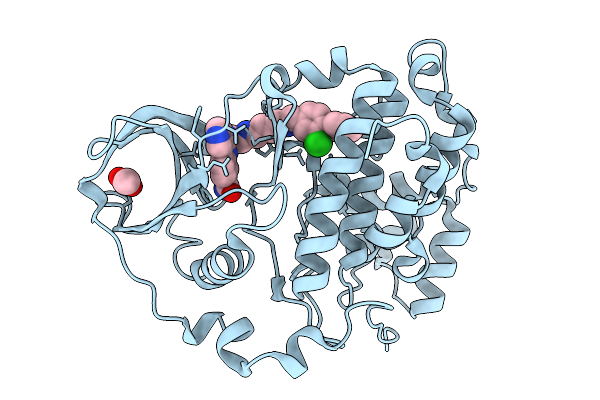 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1BZS |
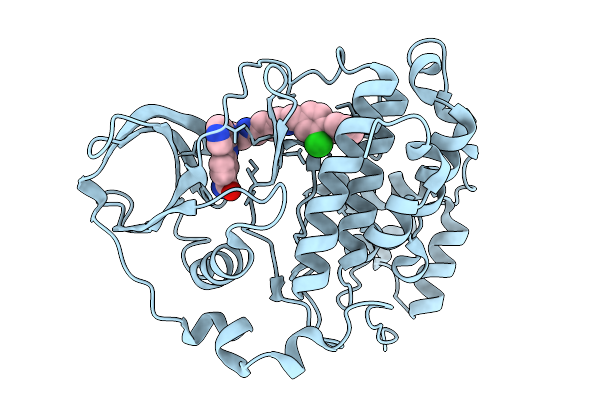 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZR |
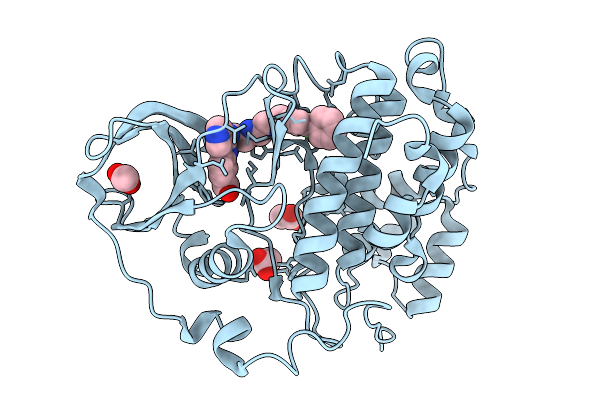 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1BZ2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZ1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZ0, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1BZZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZY, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1B0C |
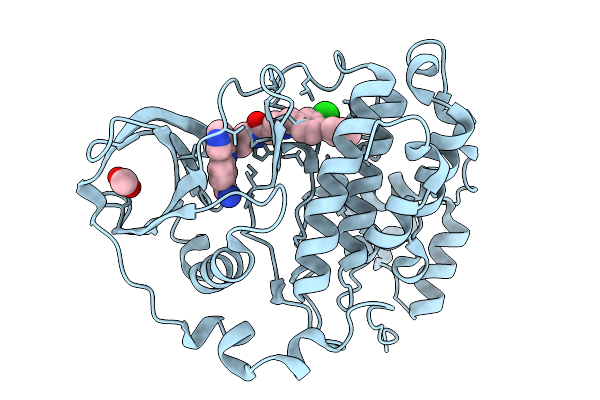 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1B0D |
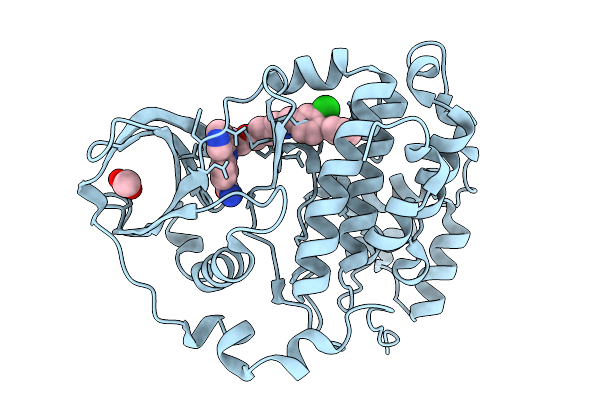 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1B0K |
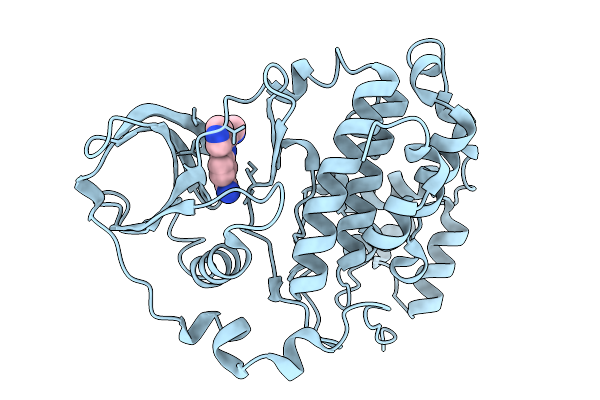 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1B0J |
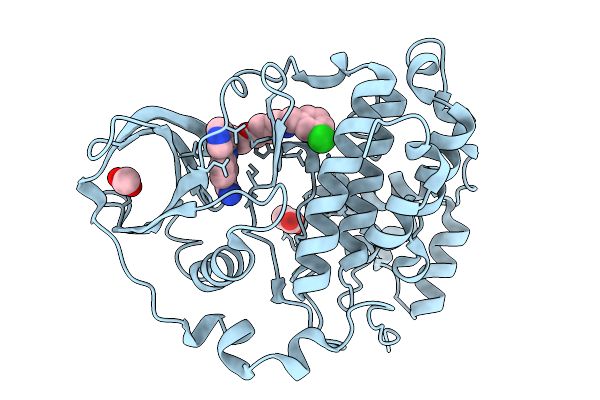 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1B0E |
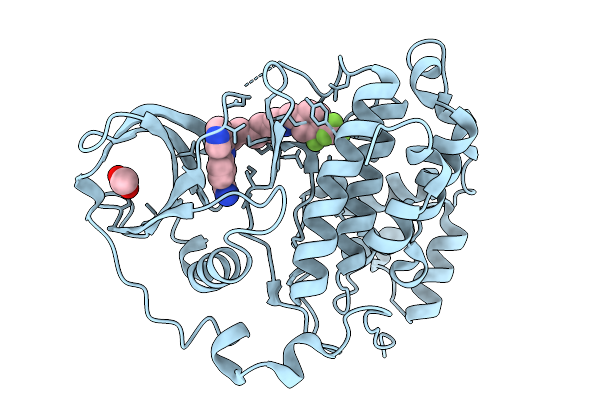 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1B0F, ACT |
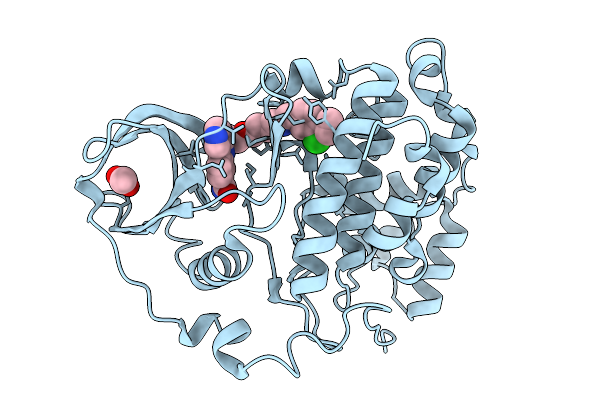 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ACT, A1B0G |
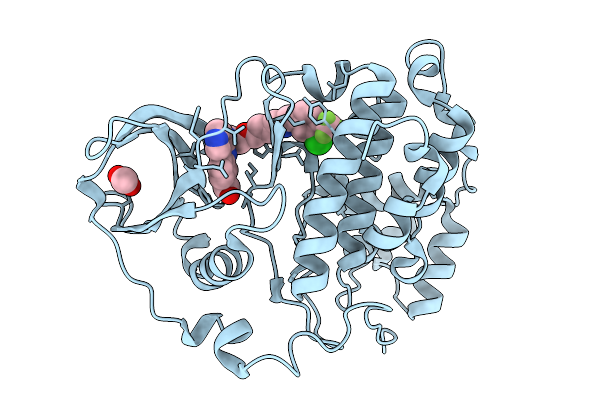 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1B0H, ACT |
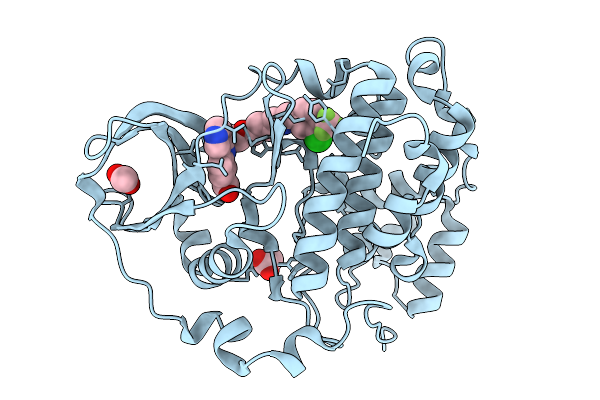 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1B0A, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZ9, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: HYDROLASE Ligands: A1BZ8, ACT |