Search Count: 92,391
 |
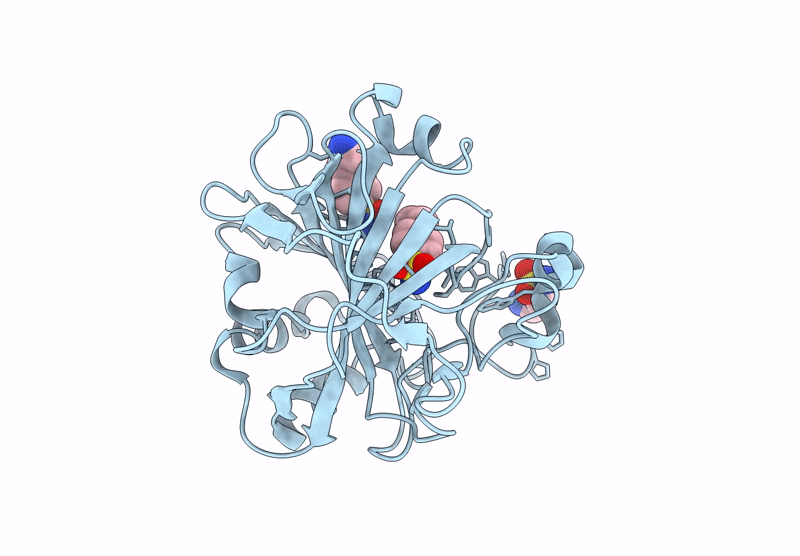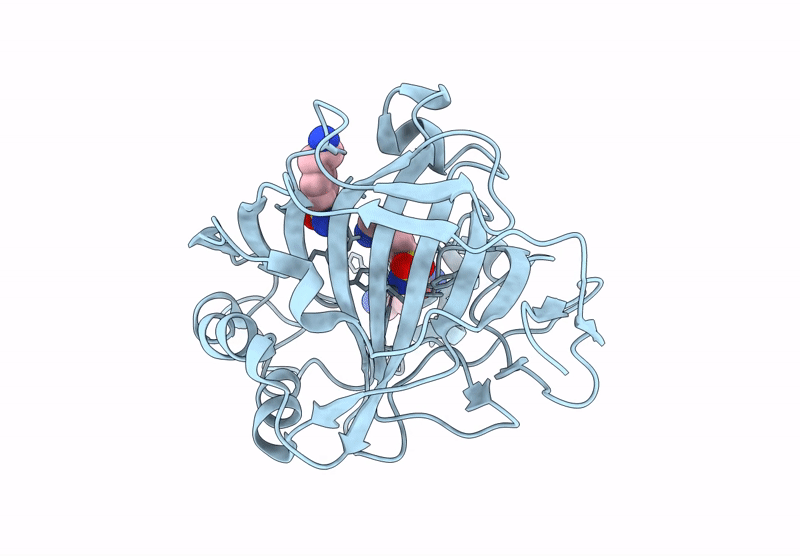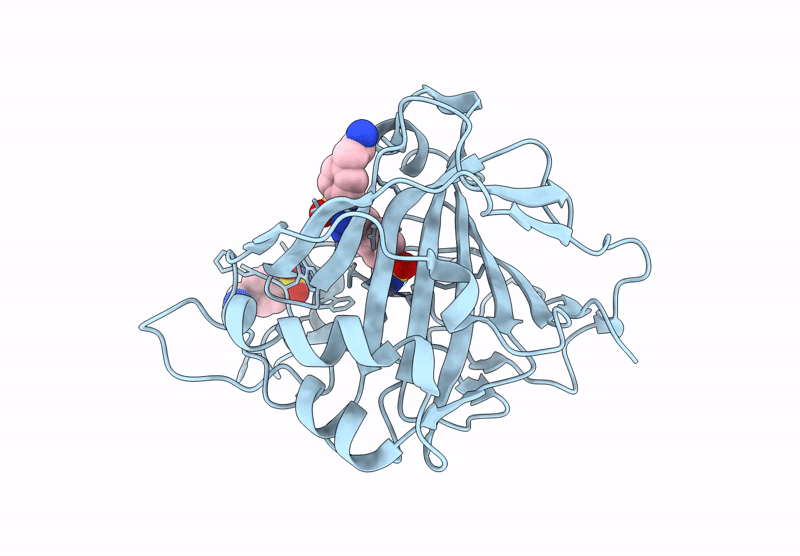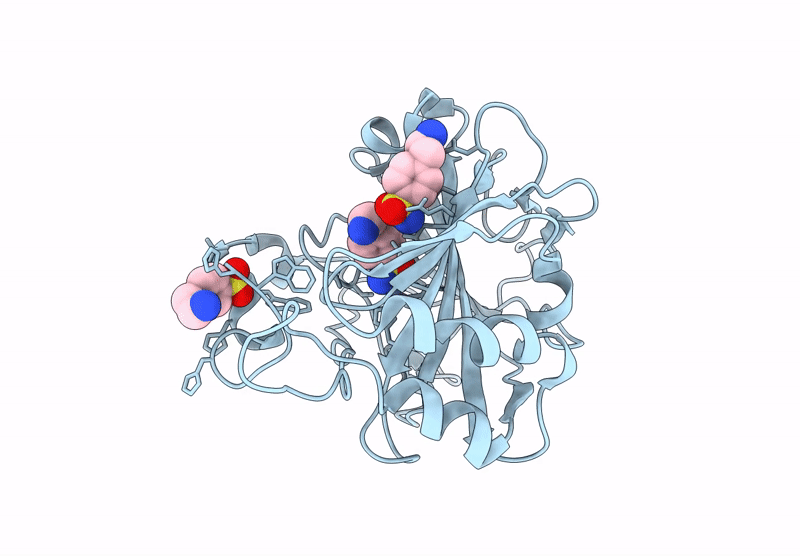
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE/INHIBITOR Ligands: ZN, A1ADH, ZYX |
 |
Hca-Ii-4-(2-Aminoethyl)Benzenesulfonamide-Complex. Two Molecules Of Inhibitor Bound To Active Site And Secondary Binding Pocket
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE/INHIBITOR Ligands: ZN, ZYX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE/INHIBITOR Ligands: ZN |
 |
Sars-Cov-2 Papain-Like-Protease (Plpro) In Complex With Inhibitor Linagliptin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: 356, GOL |
 |
Organism: Rattus norvegicus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: CLR |
 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP |
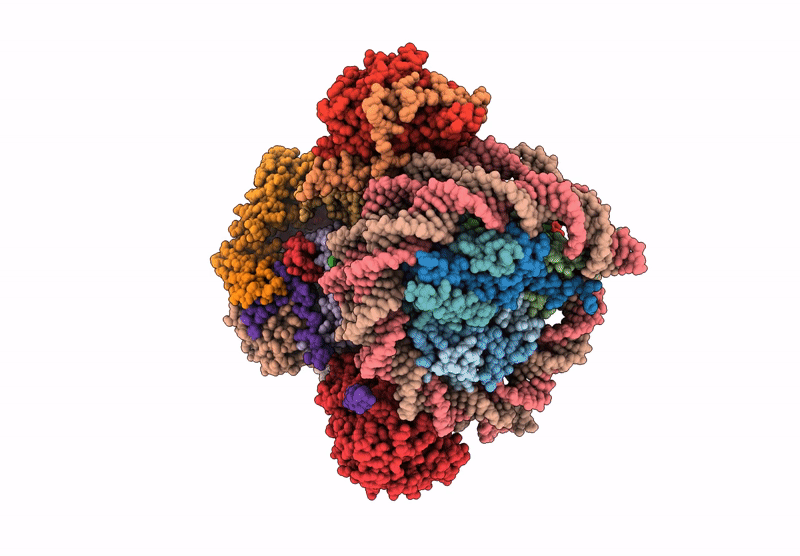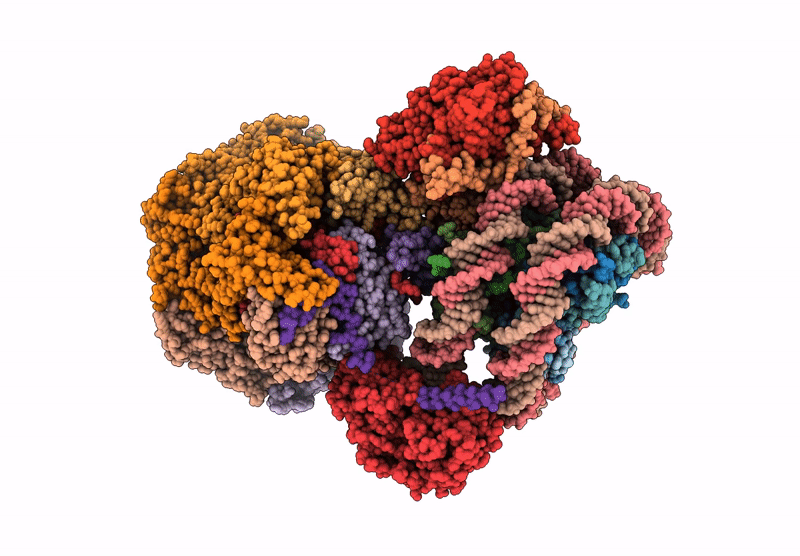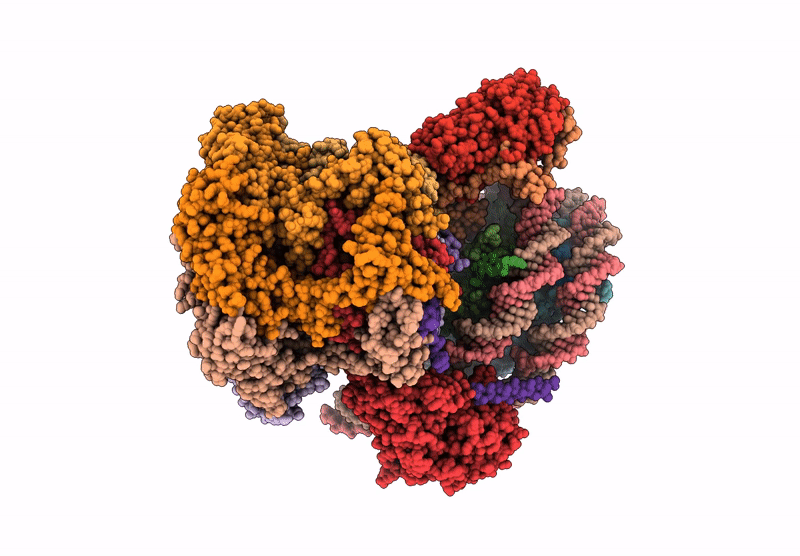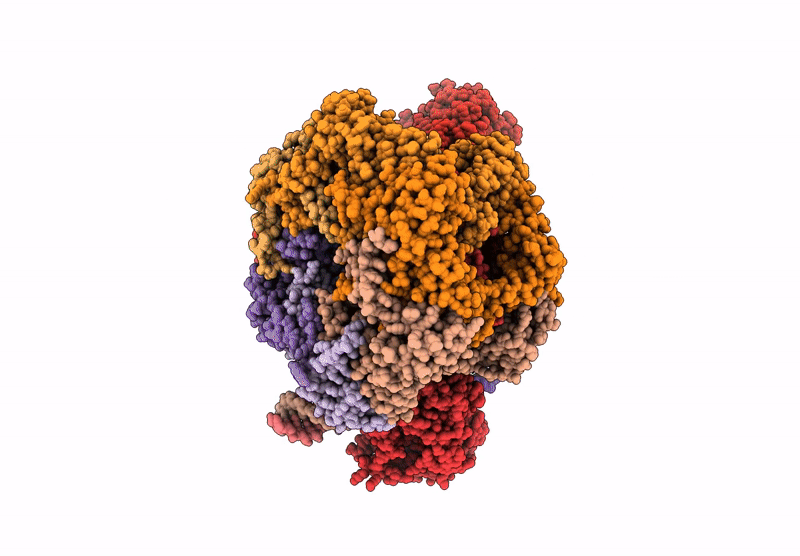 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP |
 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP |
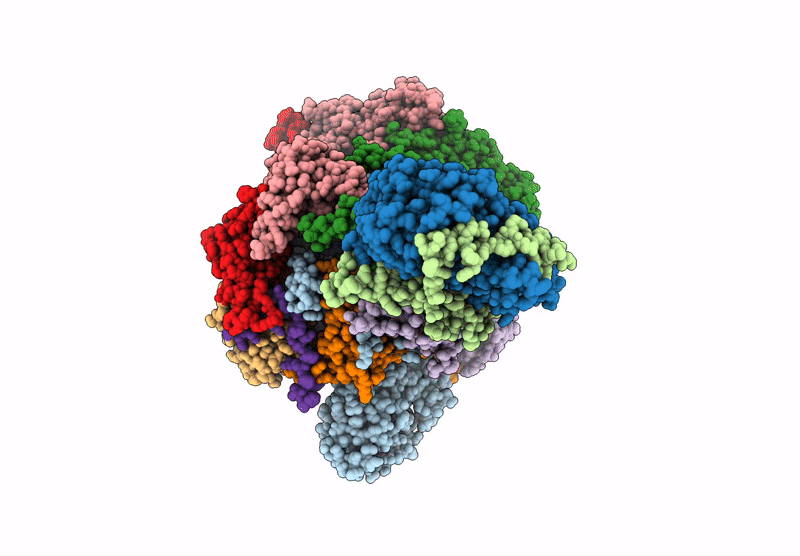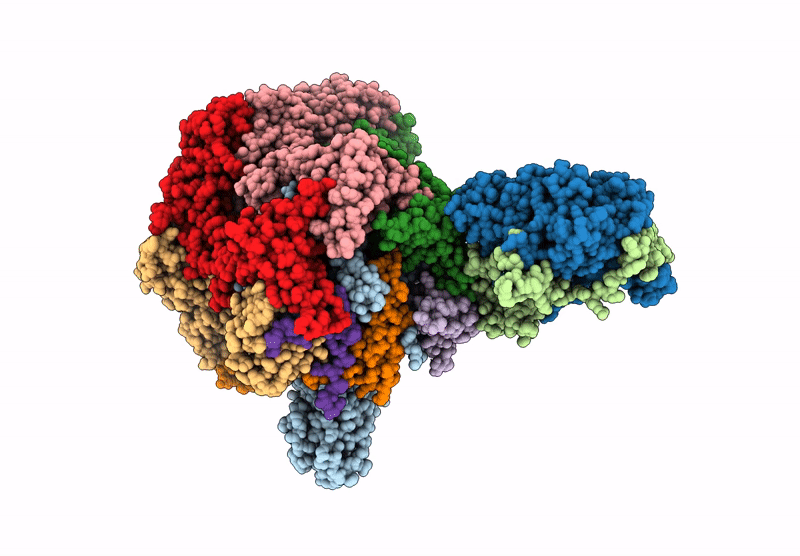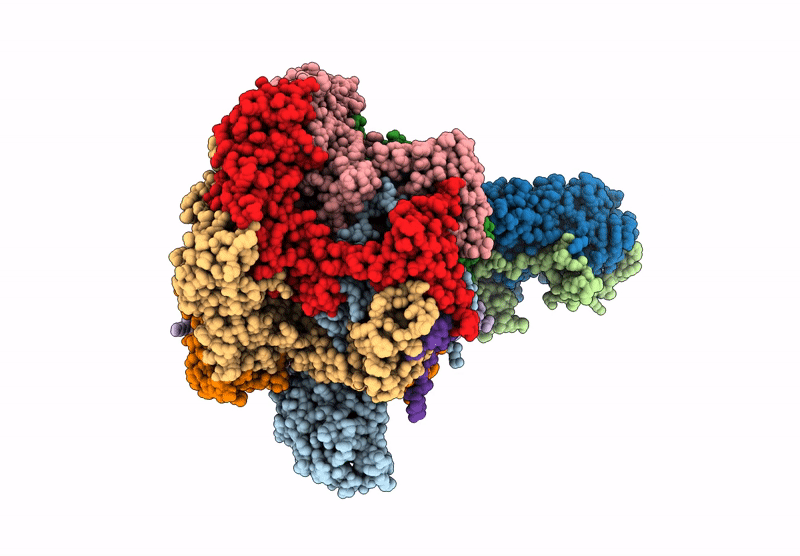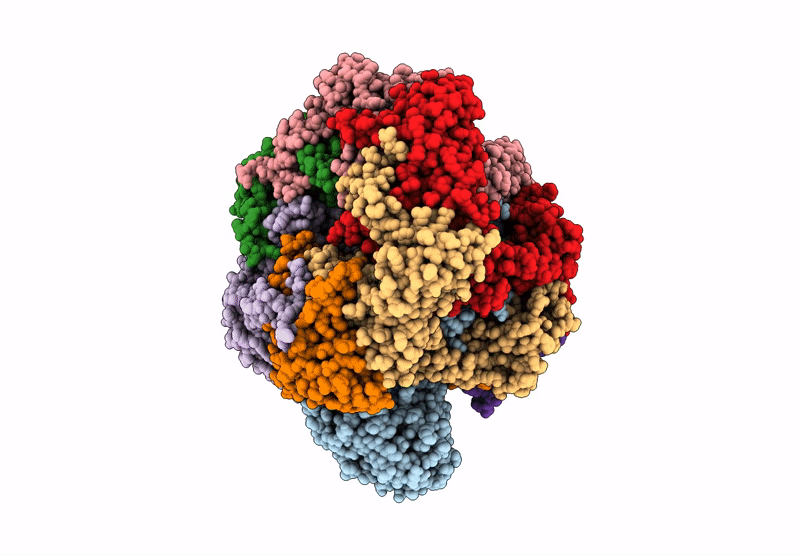 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
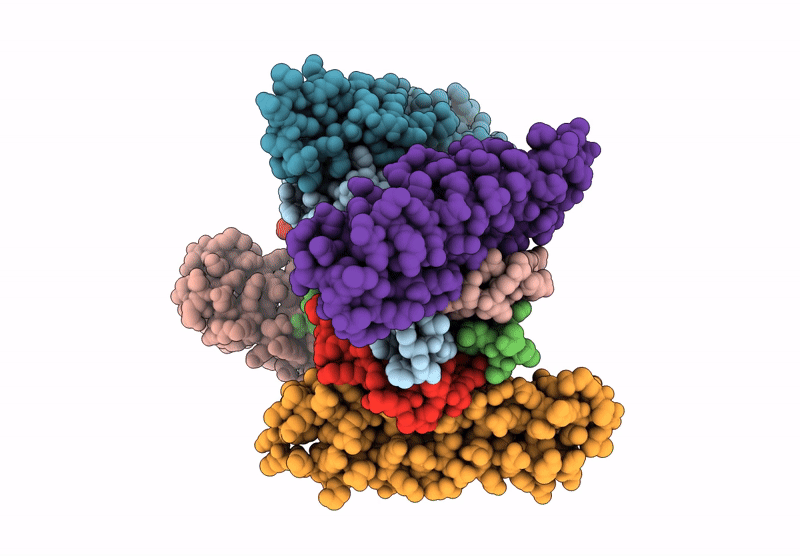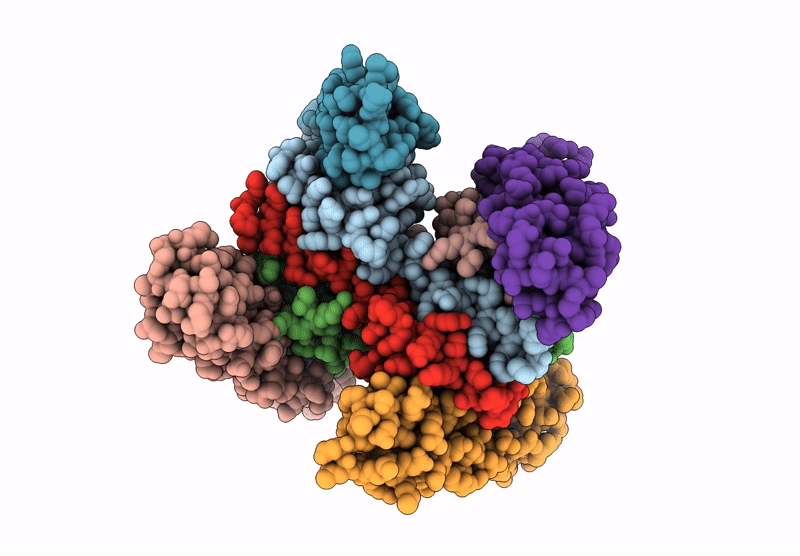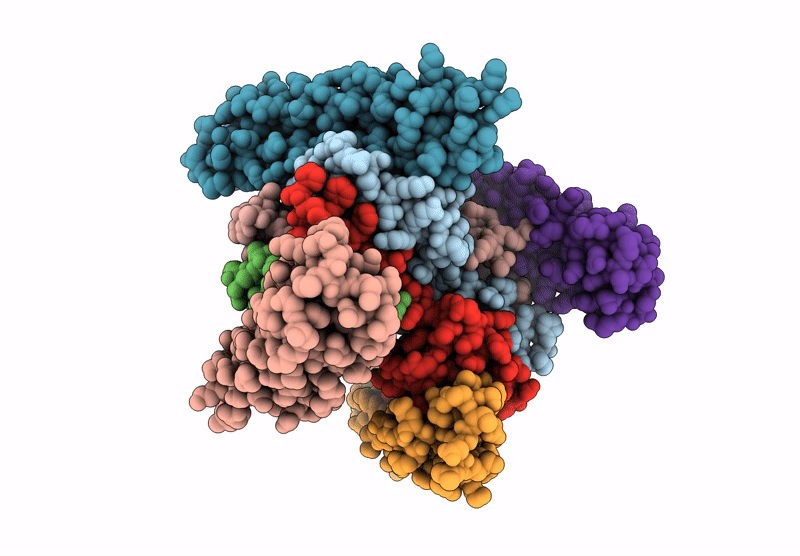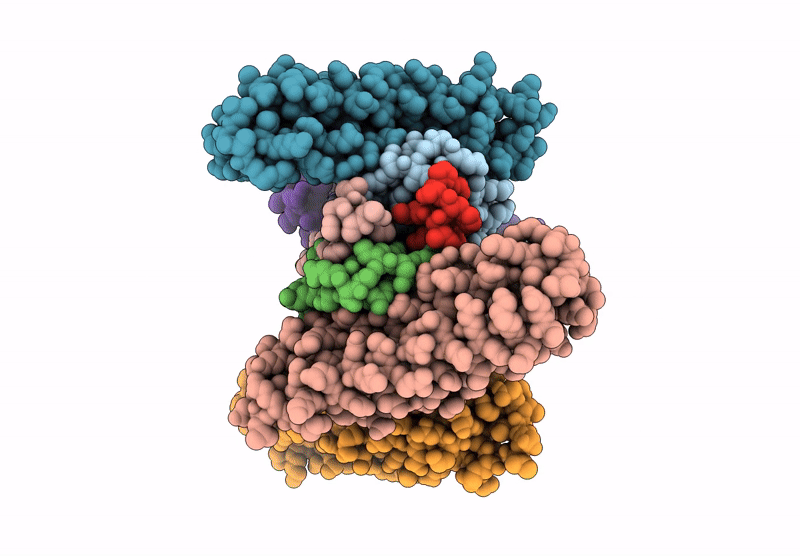
Organism: Saccharomyces cerevisiae, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: A1AZA, ACT, EDO, ANP, MG |
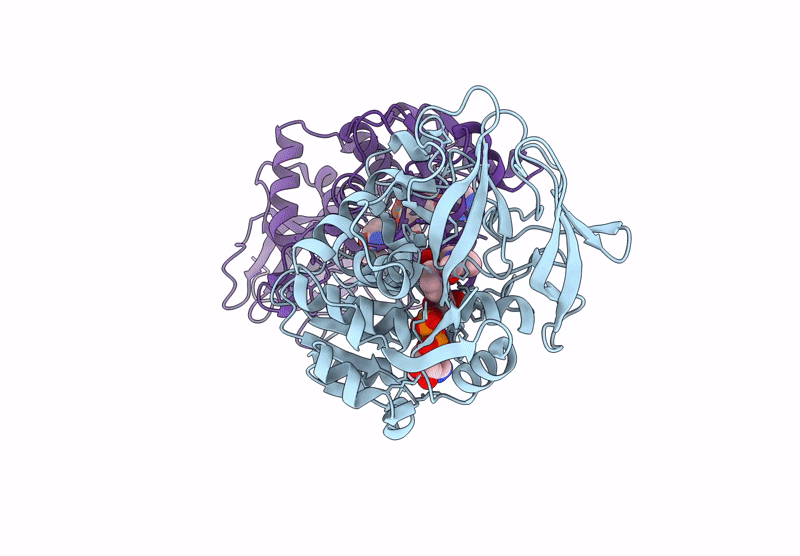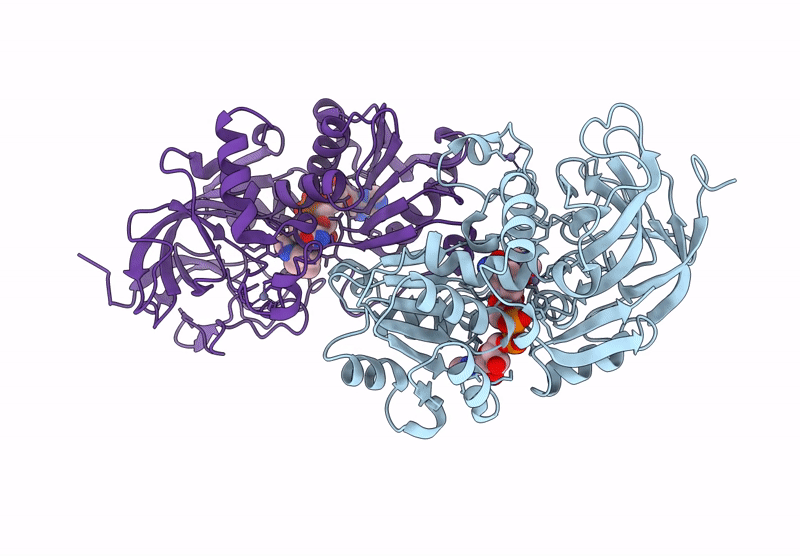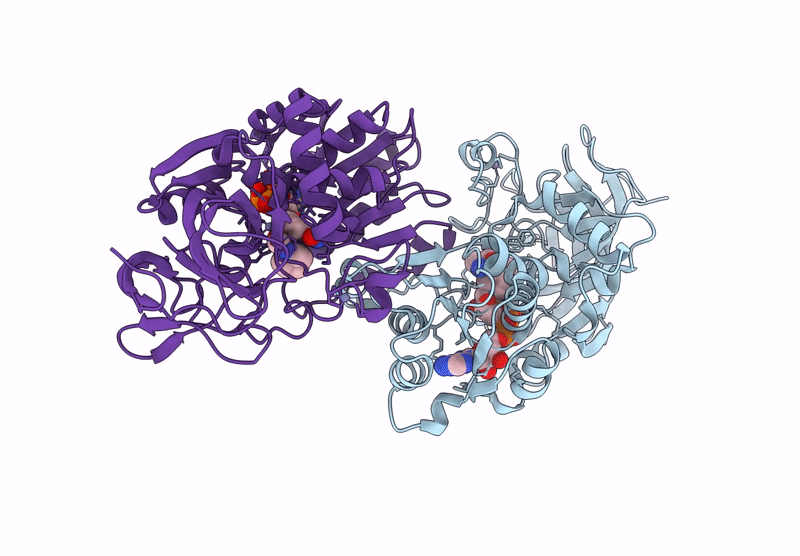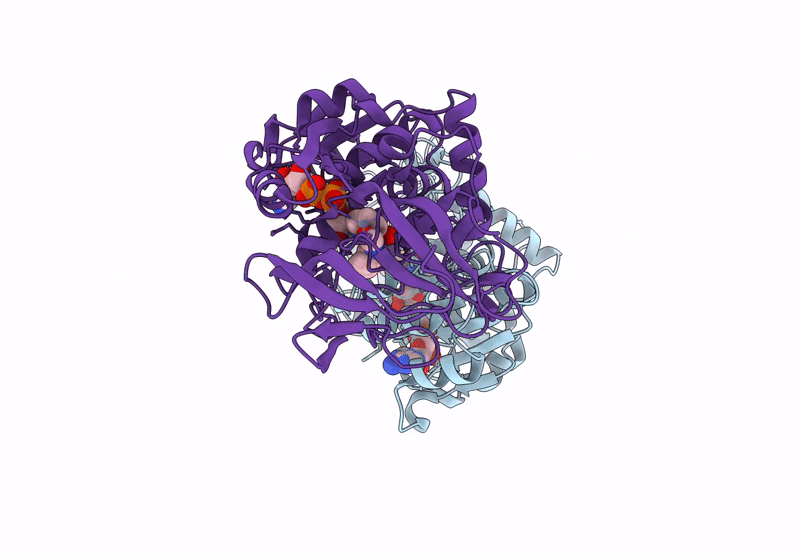 |
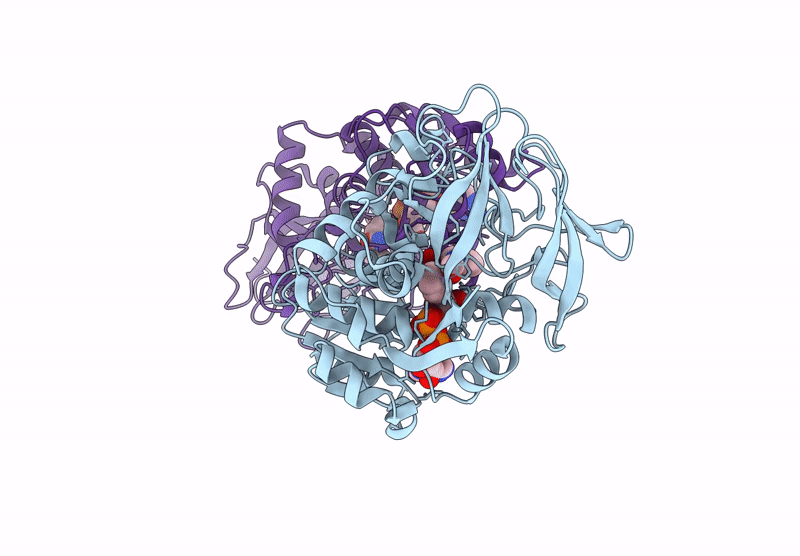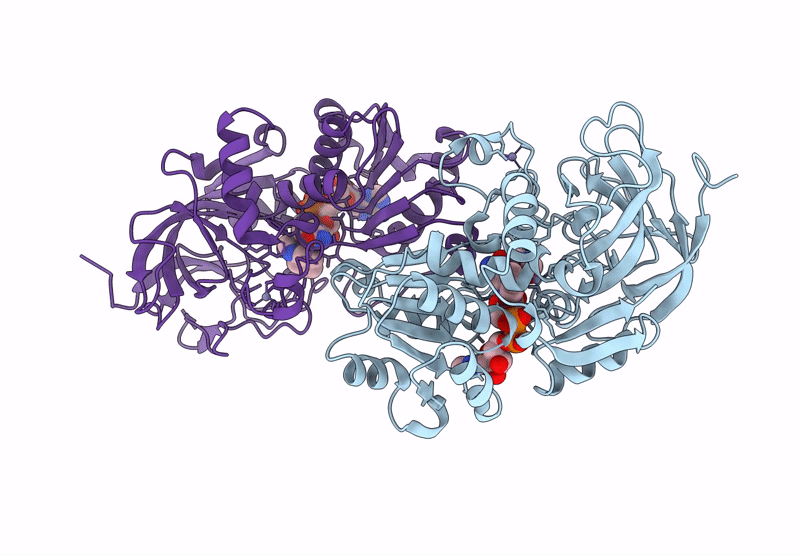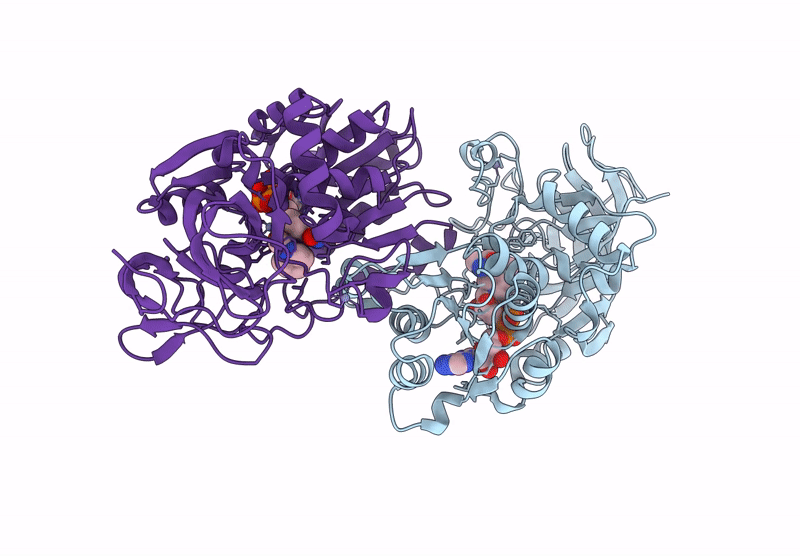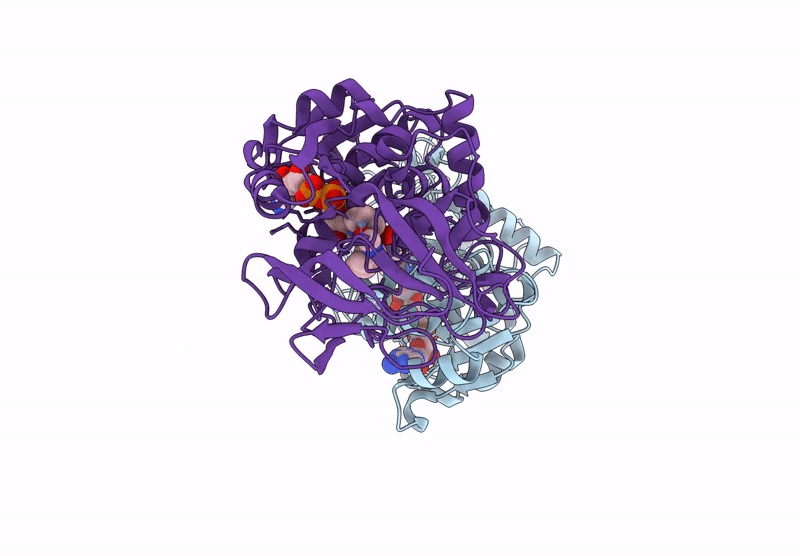
Horse Liver Alcohol Dehydrogenase V203L In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Horse Liver Alcohol Dehydrogenase V203A In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PROTEIN BINDING Ligands: TA1, GDP, GTP, MG |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PROTEIN BINDING Ligands: ADP, MG, TA1, GDP, GTP |
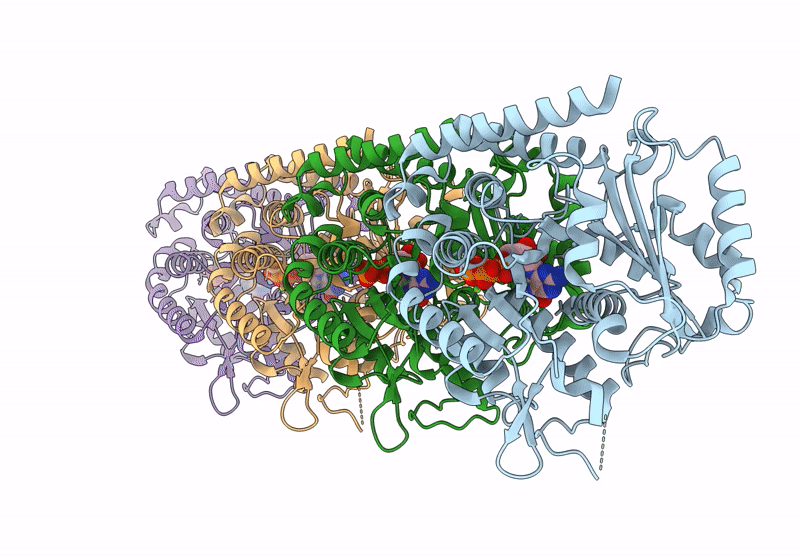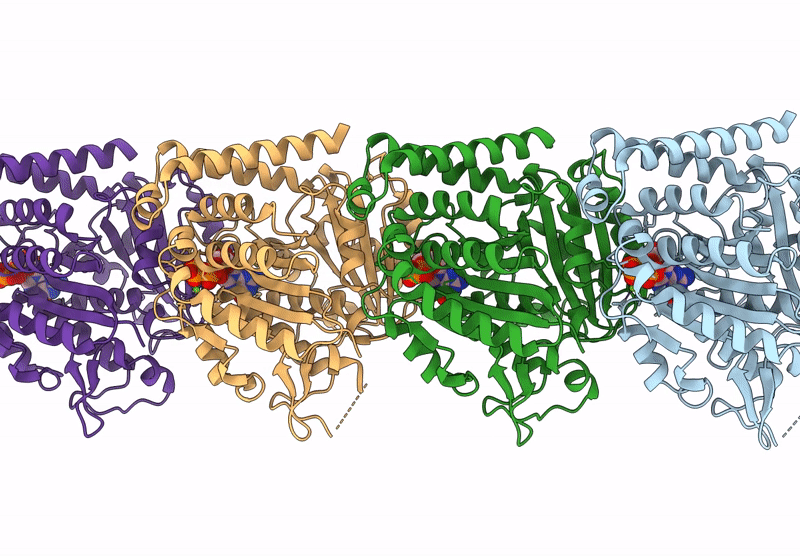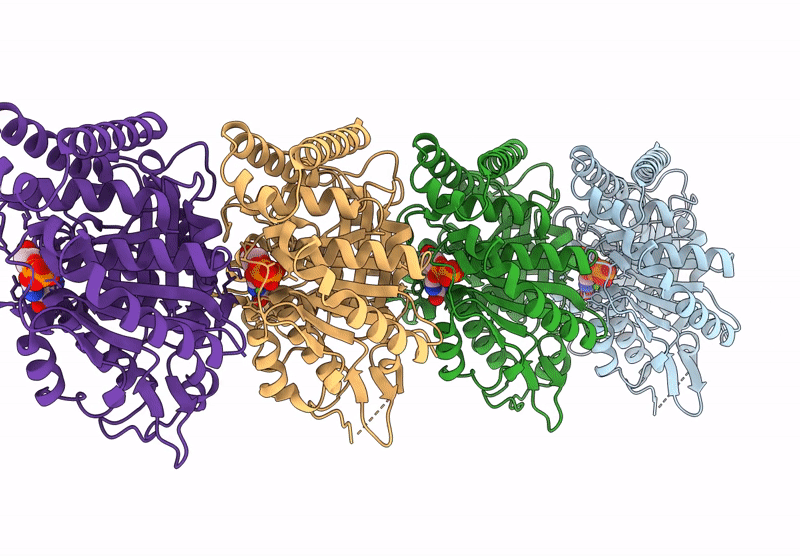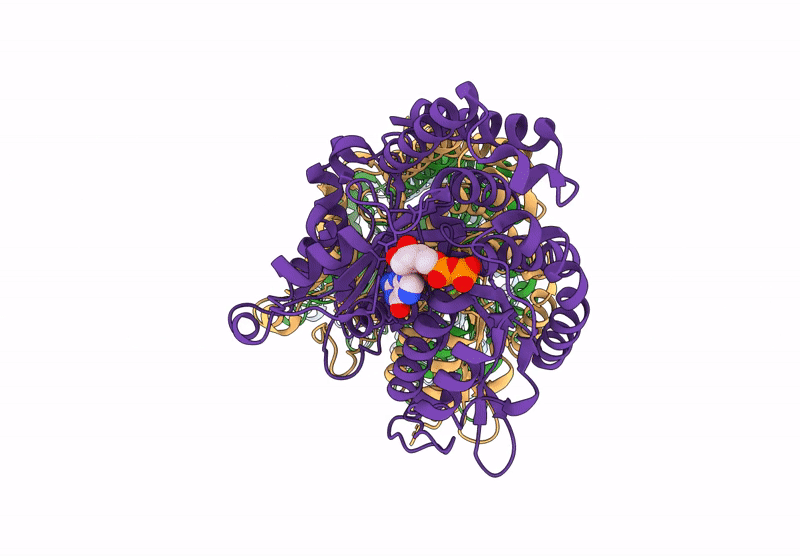 |
Model Of Tubulin Dimers Used For Determining The Dimer Rise In A Taxol-Stabilized Microtubule-Hurp Complex
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PROTEIN BINDING Ligands: GTP, MG, GDP |

