Search Count: 74,162
 |
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163-His280Ala With (S)-2-Fluoro-3-Phenylpropanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE |
 |
Positive Allosteric Modulator(Bms986187)-Bound Delta-Opioid Receptor-Gi Complex
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: A1D6B, A1D6F |
 |
Crispr-Associated Deaminase Cad1 In Ca4 Bound Form, Symmetry Expanded Dimer, Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: PTR, SO4 |
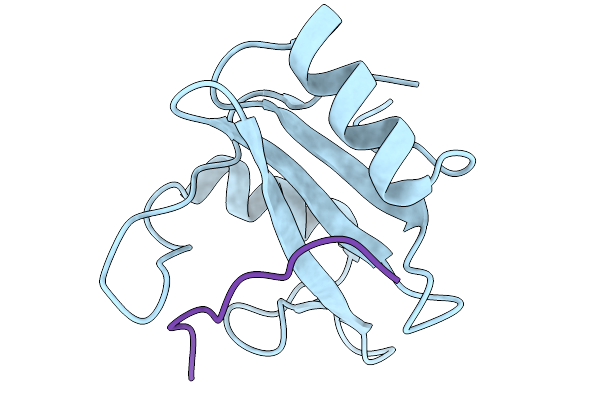 |
Crystal Structure Of N-Sh2 Domain Of Shp2 Bound To Gab1 Tyrosine Phosphorylated Peptide (624-633) Qvepyldldld
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
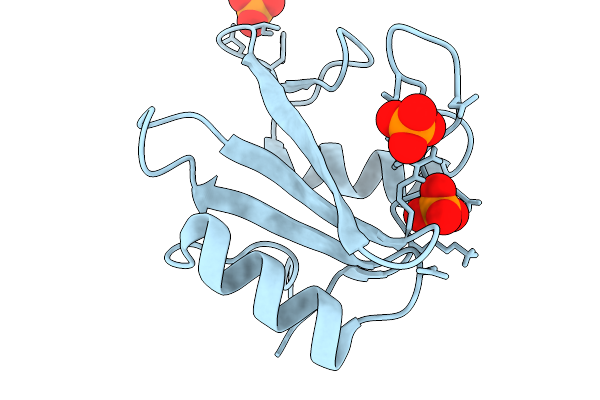 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: PO4 |
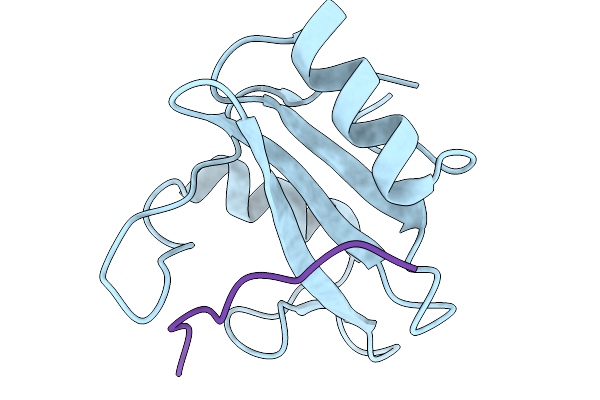 |
Crystal Structure Of N-Sh2 Domain Of Shp2 With T42A Mutation Bound To Gab1 Tyrosine Phosphorylated Peptide (624-633) Qvepyldldld
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
 |
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: GOL, SO4 |
 |
Human Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 3 (Enpp3) Inhibitor Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE Ligands: A1BYU, ZN, CA, NAG, CL |
 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Primed, Ahd 3Dva Sorted
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: CLR, EIG, GTP, MG |
 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-2
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP |
 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-3
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP, EIG |
 |
Organism: Feline infectious peritonitis virus (strain 79-1146), Felis catus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: NAG |
 |
Crystal Structure Of Pseudopedobacter Saltans Gh43 Beta-Xylosidase In Complex With Xylose.
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE Ligands: CA, XLS |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TOXIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: MES, GOL, SO4 |
 |
Crystal Structure Of Shorter Construct Of Shp2 Unbound N-Sh2 Domain (Y66 In Blocking Conformation)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
 |
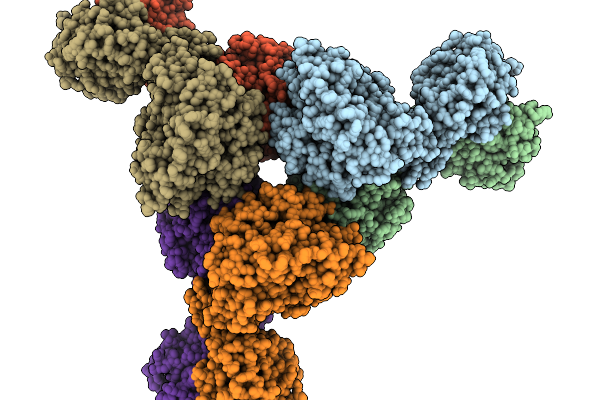
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: A1MCX, 1PE, SO4 |

