Search Count: 1,08,343
 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: A1IIY, MG |
 |
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: ELECTRON TRANSPORT Ligands: SF4, NA, FES, FMN, DU0, U10, ZN, CDL, HEM, CA, HEC, HEA, CU, MN, CUA |
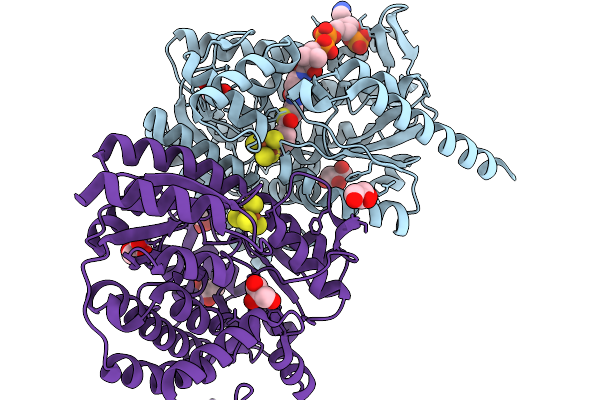 |
3-Methylbenzoyl-Coa Reductase From Thauera Chlorobenzoica (Subunits Mbdon )
Organism: Thauera chlorobenzoica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: SF4, BYC, GOL, CL |
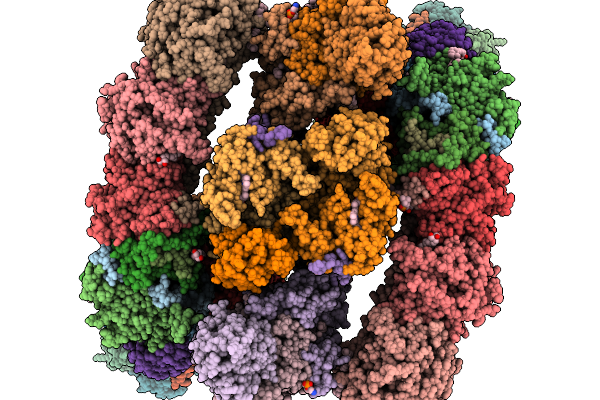 |
Respiratory Supercomplex Ci2-Ciii2-Civ2 (Megacomplex) From Alphaproteobacterium
|
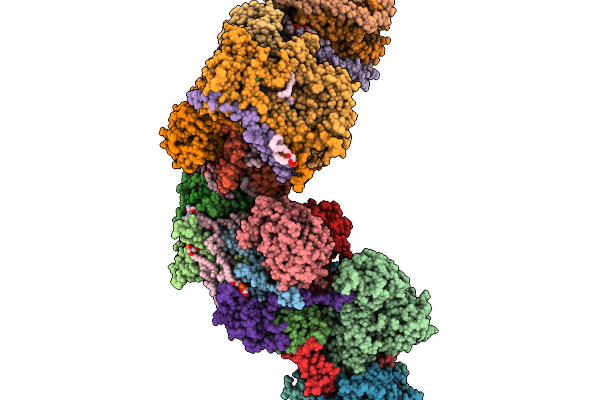 |
Respiratory Supercomplex Ci1-Ciii2-Civ1 (Respirasome) From Alphaproteobacterium
|
 |
 |
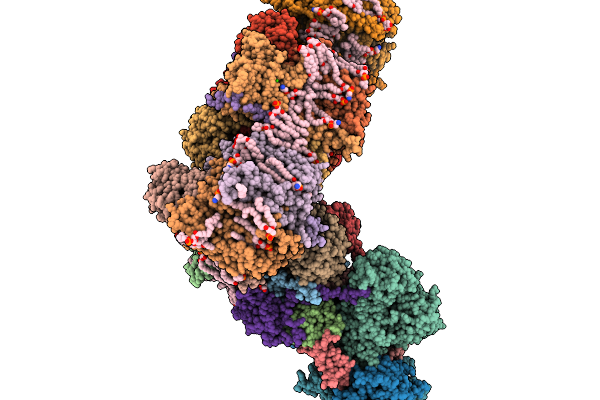
Photosynthetic A10B10 Glyceraldehyde-3-Phospahte Dehydrogenase From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Resolution:7.20 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Structure Of Carbamoylated Recombinant Human Butyrylcholinesterase By The Biscarbamte 5-(1-Hydroxy-2-(Piperidin-1-Yl)Ethyl)-1,3-Phenylene Bis(Piperidine-1-Carboxylate)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: NAG, MES, GOL, A1I0D, A1I0F, SO4 |
 |
Crystal Structure Of A Pu Hydrolysis Enzyme Aes72 From Comamonas Acidovorans
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-02-04 Classification: HYDROLASE |
 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Crystal Structure Of Glucose Tolerant Beta-Glucosidase Unbgl1 Mutant (C188V)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: CL, PO4 |
 |
Crystal Structure Of Glucsoe Bound Glucose Tolerant Gh1 Beta-Glucosidase Mutant (Unbgl1_C188V)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Crystal Structure Of Glucose Tolerant Gh1 Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, GOL, CL |
 |
Crystal Structure Of Glucose Bound Gh1 Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, GOL, PO4 |
 |
Crystal Structure Of Cellobiose And Glucose Bound Glucose Toleranant Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4, CL |
 |
High Resolution Crystal Strucutre Of Highly Glucose Tolerant Gh1 Beta-Glucosidase (Unbgl1_C188V_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of Glucose Bound Highly Glucose Tolerant Gh1 Beta-Glcosidase Mutant (Unbgl1_C188V_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4 |
 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: GS1, PO4, NA |

