Search Count: 156
 |
Crystal Structure Of The Peptidoglycan O-Acetyltransferase B (Patb) From Campylobacter Jejuni, Catalytic Domain
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: CL |
 |
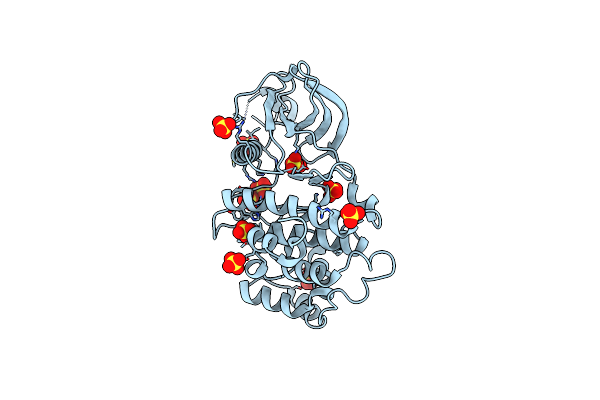
Crystal Structure Of The Levansucrase Beta From Pseudomonas Syringae Pv. Actinidiae
Organism: Pseudomonas syringae pv. actinidiae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-09-25 Classification: TRANSFERASE Ligands: PO4, P4K, ACT, GOL, SCN, MES |
 |
Crystal Structure Of Cmgc Family Protein Kinase From Trichomonas Vaginalis (Apo)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Beijerinckia indica subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: GOL |
 |
Beijerinckia Indica Beta-Fructosyltransferase Variant H395R/F473Y In Complex With Fructose
Organism: Beijerinckia indica subsp. indica nbrc 3744
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: MG, FRU, BDF |
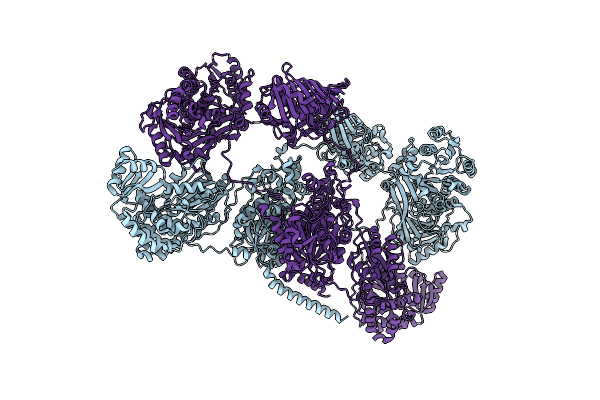 |
Organism: Streptomyces chartreusis nrrl 3882
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: TRANSFERASE |
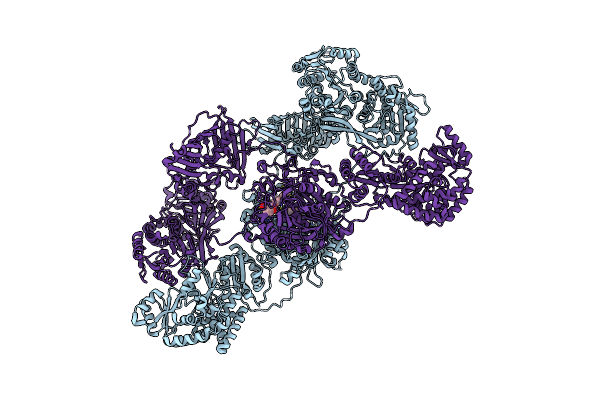 |
Organism: Streptomyces chartreusis nrrl 3882
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: TRANSFERASE Ligands: ONF, 3HA |
 |
Crystal Structure Of Wild-Type From Arabidopsis Thaliana Complexed With Galactose
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-11-30 Classification: TRANSFERASE Ligands: GAL |
 |
Crystal Structure Of D383A Mutant From Arabidopsis Thaliana Complexed With Galactose.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-11-30 Classification: TRANSFERASE Ligands: GAL |
 |
Crystal Structure Of D383A Mutant From Arabidopsis Thaliana Complexed With Galactinol.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2022-11-30 Classification: TRANSFERASE Ligands: BQZ |
 |
Crystal Structure Of Alkaline Alpha-Galctosidase D383A Mutant From Arabidopsis Thaliana Complexed With Raffinose
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-11-30 Classification: TRANSFERASE |
 |
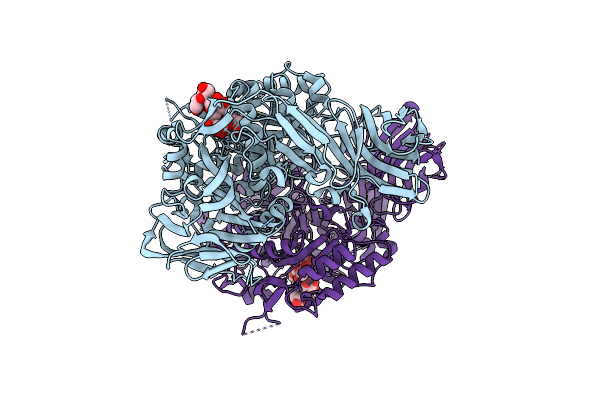
Crystal Structure Of Alkaline Alpha-Galactosidase D383A Mutant From Arabidopsis Thaliana Complexed With Product-Galactose And Sucrose.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-11-30 Classification: TRANSFERASE Ligands: GAL |
 |
Crystal Structure Of Alkaline Alpha-Galactosidase D383A Mutant From Arabidopsis Thaliana Complexed With Stachyose.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-30 Classification: TRANSFERASE |
 |
Crystal Structure Of 3-Deoxy-Manno-Octulosonate Cytidylyltransferase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae hs11286
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-09-14 Classification: TRANSFERASE |
 |
Crystal Structure Of Haloarcula Marismortui Cheb With Glutathione S-Transferase Expression Tag
Organism: Schistosoma japonicum, Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-07-20 Classification: TRANSFERASE, HYDROLASE |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With (E)-3-(3-Chloro-5-(2-(2-(2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Ethoxy)-4-(2-Morpholinoethoxy)Phenoxy)Phenyl)Acrylonitrile (Jlj530)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9UV |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With 6-((2-((4-Cyanophenyl)Amino)Pyrimidin-4-Yl)Amino)-5,7-Dimethylindolizine-2-Carbonitrile (Jlj604)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9WI, SO4, MG, PUT |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1, Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-03-16 Classification: HYDROLASE, Transferase/Viral Protein Ligands: 9W3 |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase Y181C Variant In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2022-03-16 Classification: hydrolase, transferase/viral protein Ligands: 9W3 |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase K103N/Y181C Variant In Complex With (E)-4-((4-((4-(2-Cyanovinyl)-2,6-Dimethylphenyl)Amino)-6-(3-Morpholinopropoxy)-1,3,5-Triazin-2-Yl)Amino)Benzonitrile (Jlj564)
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10)
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2022-03-16 Classification: hydrolase, transferase/viral protein Ligands: 9W3, MG |

