Search Count: 285
 |
Organism: Streptococcus thermophilus, Streptococcus phage 2972
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Streptococcus thermophilus, Streptococcus phage vb_sths-va214
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN |
 |
Organism: Francisella tularensis subsp. novicida
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: STRUCTURAL PROTEIN |
 |
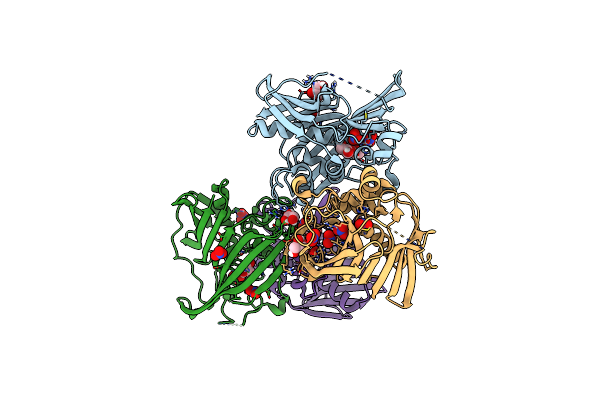
Organism: Chlamydia trachomatis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-03-13 Classification: HYDROLASE Ligands: PO4, MG, GOL, PEG, PGE |
 |
Organism: Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-12-27 Classification: UNKNOWN FUNCTION Ligands: GOL, NO3 |
 |
Major Interface Of Streptococcal Surface Enolase Dimer From Ap53 Group A Streptococcus Bound To A Lipid Vesicle
Organism: Streptococcus pyogenes
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: LYASE |
 |
Major Interface Of Streptococcal Surface Enolase Dimer From Ap53 Group A Streptococcus Bound To A Lipid Vesicle
Organism: Streptococcus pyogenes
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: LYASE |
 |
Organism: Streptococcus pyogenes
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: LYASE |
 |
Organism: Streptococcus sp. 'group a'
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: LYASE |
 |
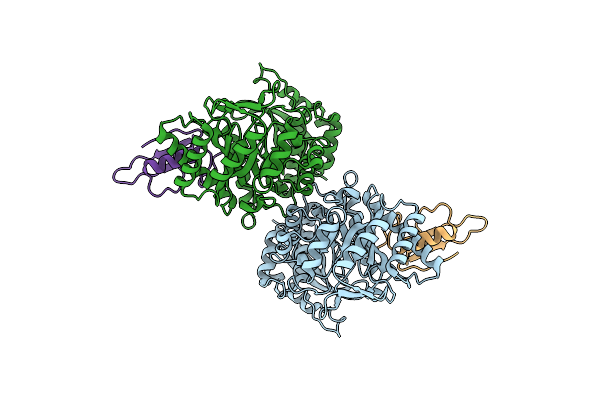
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-11-09 Classification: LYASE Ligands: PO4, MG, ACT, EDO, PGE, SEP |
 |
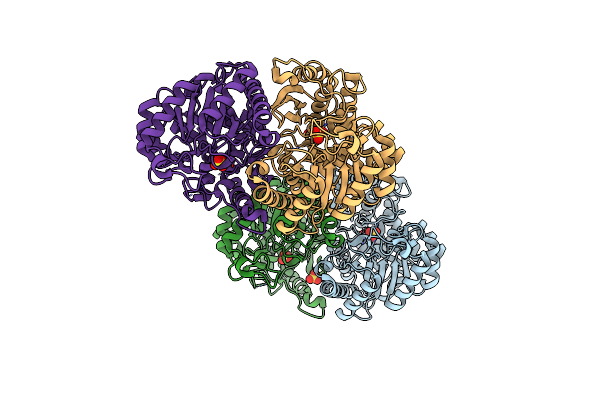
Organism: Bacillus subtilis (strain 168), Bacillus phage sp01
Method: ELECTRON MICROSCOPY Release Date: 2022-07-27 Classification: LYASE/LYASE INHIBITOR Ligands: MG |
 |
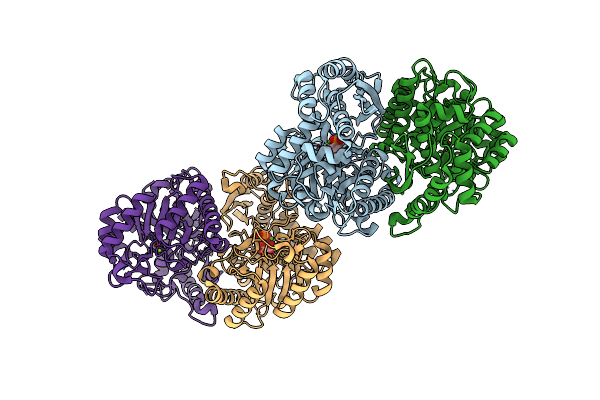
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: METAL BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Enolase1 From Candida Albicans Complexed With 2'-Phosphoglyceric Acid Sodium
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-06 Classification: METAL BINDING PROTEIN Ligands: MG, 2PG |
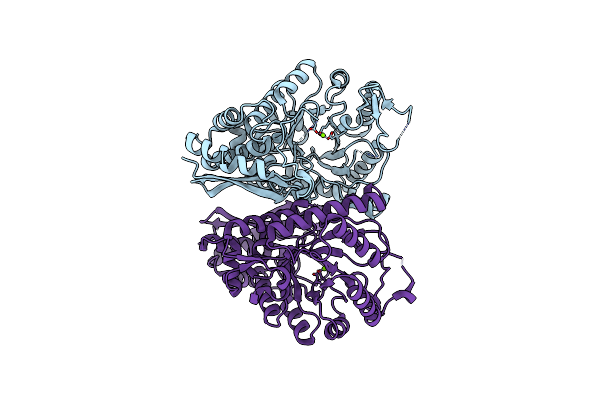 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-16 Classification: LYASE Ligands: MG |
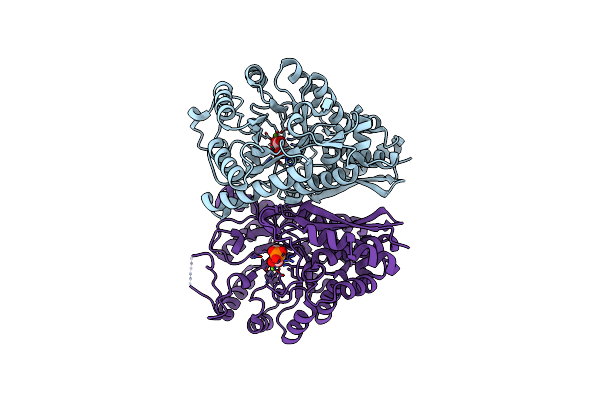 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-03-16 Classification: LYASE Ligands: 2PG, MG |
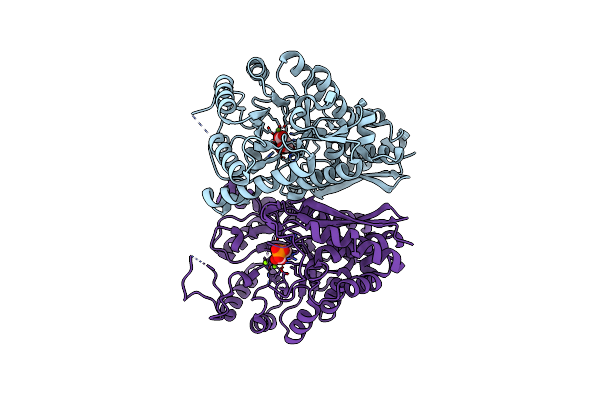 |
Aspergillus Fumigatus Enolase Bound To Phosphoenolpyruvate And 2-Phosphoglycerate
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-16 Classification: LYASE Ligands: MG, 2PG, PEP |
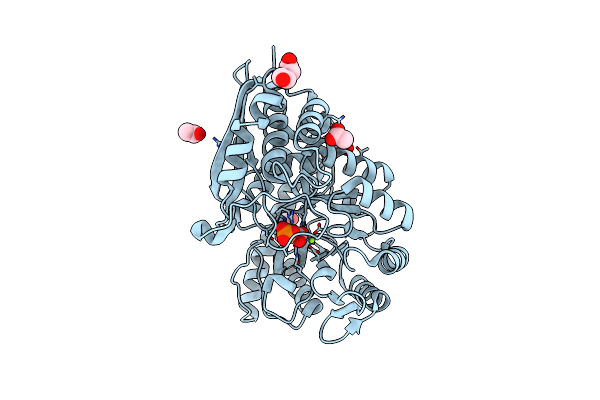 |
Mycobacterium Tuberculosis Enolase Mutant - E204A Complex With Phosphoenolpyruvate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: PEP, PEG, EDO, ACT, MG |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN Ligands: PEP, MG |
 |
Organism: Mycoplasma bovis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-02-02 Classification: LYASE |

