Search Count: 180
All
Selected
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-26 Classification: HYDROLASE Ligands: CA, EDO |
 |
Crystal Structure Of Zmmoc1/Nicked Holliday Junction Complex At Ground State
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-06-26 Classification: HYDROLASE |
 |
Crystal Structure Of Zmmoc1 In Complex With A Nicked Holliday Junction Soaked In Mn2+ For 15 Seconds
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-06-26 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Zmmoc1 In Complex With A Nicked Holliday Junction Soaked In Mn2+ For 180 Seconds
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: HYDROLASE Ligands: MN, EDO |
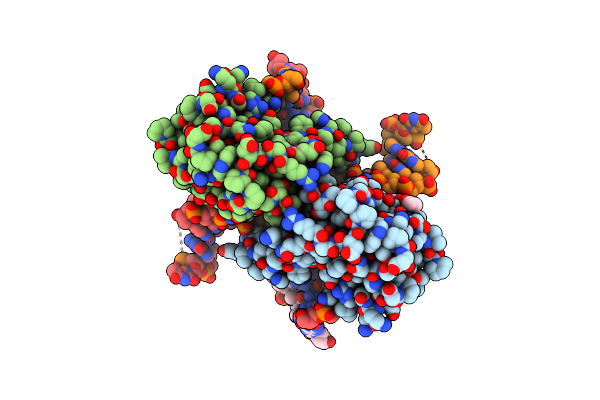 |
Crystal Structure Of Zmmoc1 K229A In Complex With A Nicked Holliday Junction Soaked In Mn2+ For 180 Seconds
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2024-06-26 Classification: HYDROLASE Ligands: MN, EDO |
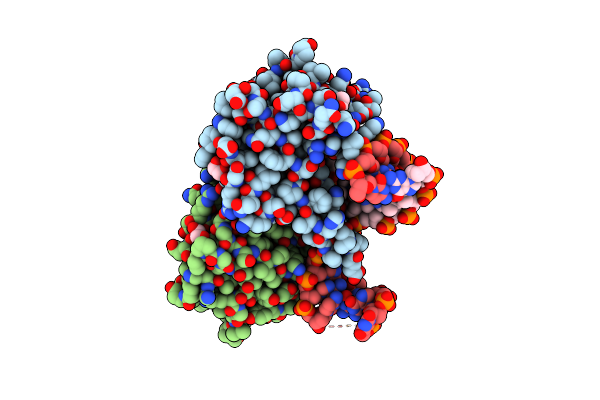 |
Crystal Structure Of Zmmoc1 K229A In Complex With A Nicked Holliday Junction Soaked In Mn2+ For 600 Seconds
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: DNA BINDING PROTEIN Ligands: MN, EDO |
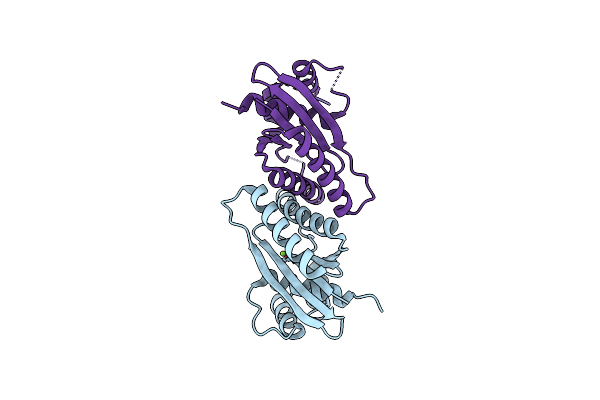 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-08-17 Classification: HYDROLASE Ligands: MG |
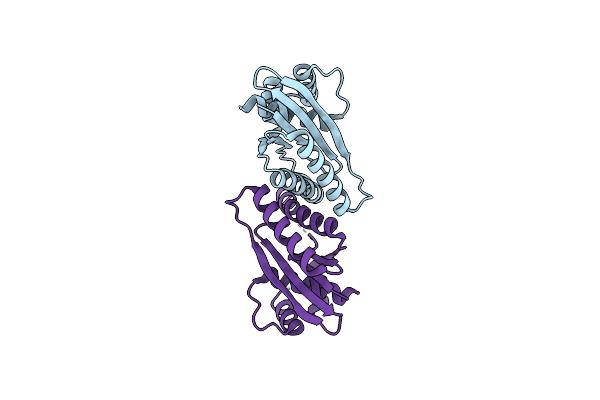 |
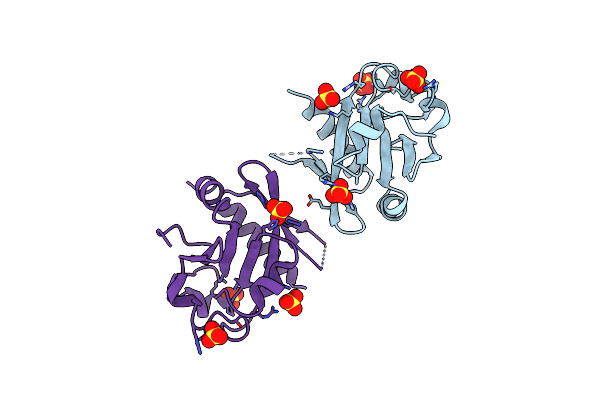
Organism: Deinococcus radiodurans atcc 13939
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-06 Classification: DNA BINDING PROTEIN |
 |
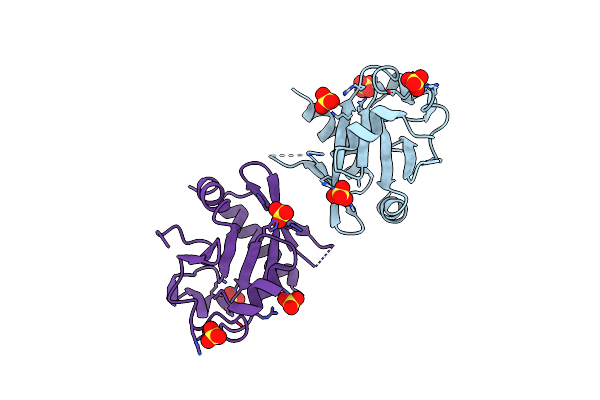
Organism: Thermus thermophilus phage 15-6
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-02-16 Classification: RECOMBINATION Ligands: SO4 |
 |
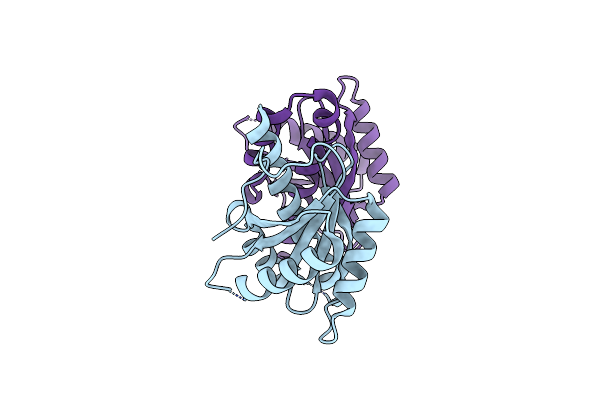
Organism: Thermus thermophilus phage 15-6
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-19 Classification: RECOMBINATION Ligands: SO4 |
 |
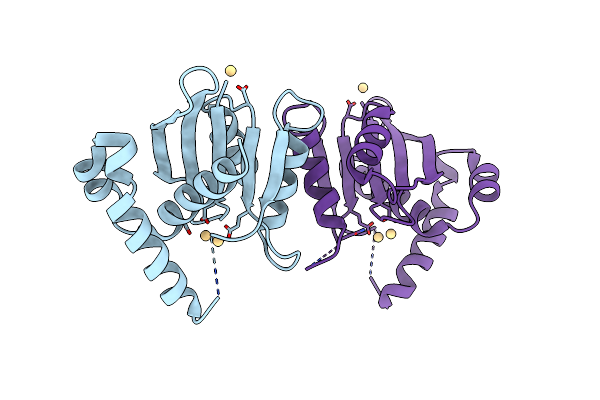
Structure-Guided Studies Of The Holliday Junction Resolvase Ruvx Provide Novel Insights Into Atp-Stimulated Cleavage Of Branched Dna And Rna Substrates
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-05-26 Classification: DNA BINDING PROTEIN |
 |
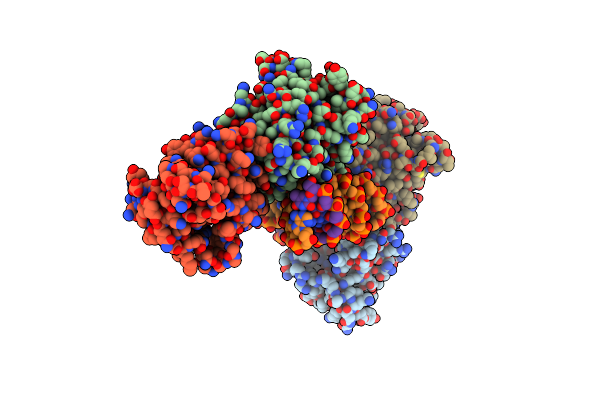
Organism: Fowlpox virus
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: CD |
 |
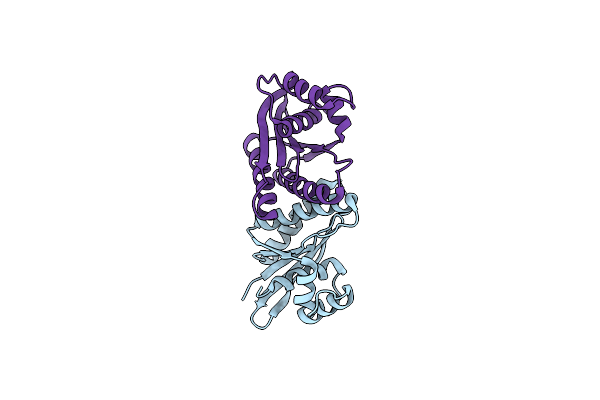
Crystal Structure Of Fowlpox Virus Resolvase And Substrate Holliday Junction Dna Complex
Organism: Fowlpox virus
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2020-04-29 Classification: HYDROLASE/DNA |
 |
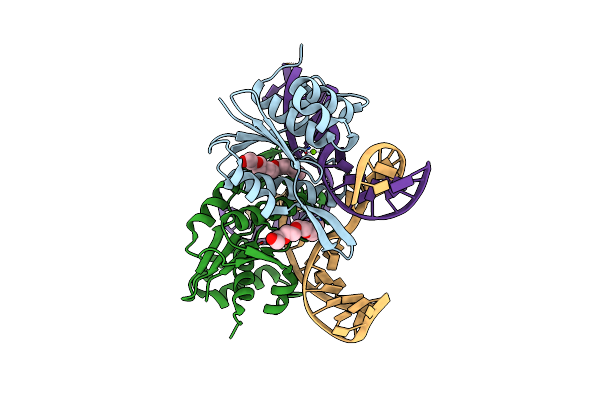
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-02-26 Classification: DNA BINDING PROTEIN |
 |
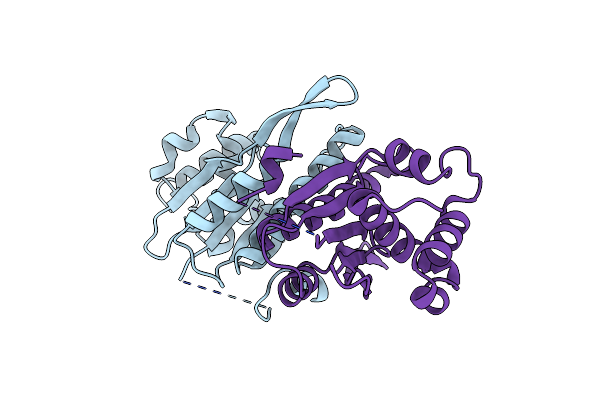
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2019-10-23 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, 1PE |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-10-23 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Zmmoc1-Holliday Junction Complex In The Presence Of Calcium
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-10-23 Classification: DNA BINDING PROTEIN/DNA Ligands: CA, 1PE |
 |
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-10-23 Classification: DNA BINDING PROTEIN/DNA Ligands: MG |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2019-09-25 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2016-12-07 Classification: HYDROLASE |

