Search Count: 274
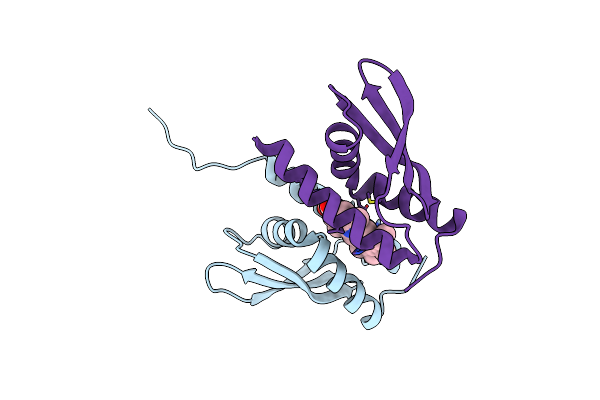 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: HEB |
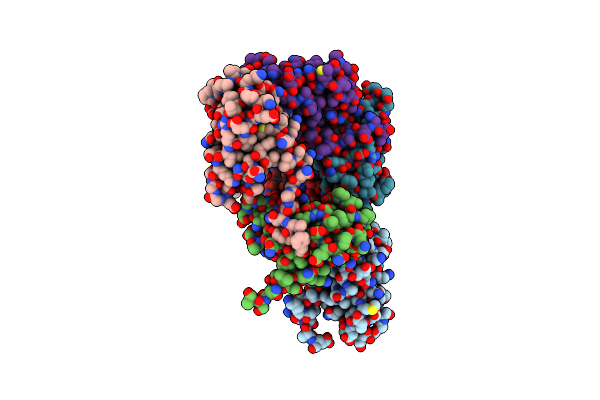 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: HEB |
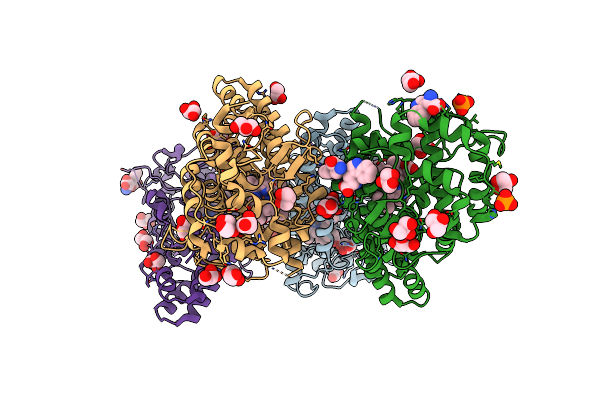 |
Crystal Structure Of L-Trp/Indoleamine 2,3-Dioxygenagse 1 (Hido1) Complex With The Jk-Loop Refined In The Closed Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: TRP, NFK, GOL, HEM, NA, PO4, CL |
 |
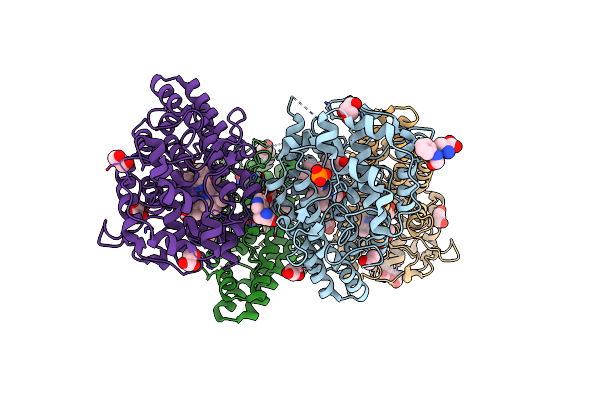
Crystal Structure Of L-Trp/Indoleamine 2,3-Dioxygenagse 1 (Hido1) Complex With The Jk-Loop Refined In The Open Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: HEM, NFK, PO4, GOL, OXY, TRP, CL |
 |
Crystal Structure Of L-Trp/Indoleamine 2,3-Dioxygenase (Hido1) Complex With The Jk-Loop Refined In The Intermediate Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, NFK, PO4, TRP |
 |
Solution Structure Of The Iwp-051-Bound H-Nox From Shewanella Woodyi In The Fe(Ii)Co Ligation State
Organism: Shewanella woodyi
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: SIGNALING PROTEIN Ligands: IWP, HEM, CMO |
 |
Solution Structure Of The H-Nox Protein From Shewanella Woodyi In The Fe(Ii)Co Ligation State
Organism: Shewanella woodyi
Method: SOLUTION NMR Release Date: 2020-05-06 Classification: SIGNALING PROTEIN Ligands: CMO, HEM |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-06-13 Classification: METAL BINDING PROTEIN Ligands: PO4, K, SF4, SRM |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-06-13 Classification: METAL BINDING PROTEIN Ligands: PO4, K, SF4, SRM |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-06-13 Classification: METAL BINDING PROTEIN Ligands: PO4, K, SF4, SRM |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-06-13 Classification: METAL BINDING PROTEIN Ligands: PO4, K, SF4, SRM |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-09 Classification: ELECTRON TRANSPORT Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2018-03-14 Classification: ELECTRON TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-14 Classification: ELECTRON TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-03-14 Classification: ELECTRON TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2017-11-22 Classification: ELECTRON TRANSPORT |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-03 Classification: HEME BINDING PROTEIN Ligands: HEB, CL, EDO |
 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii) Ligation State, Q154A/Q155A/K156A Mutant
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, ZN, NA |
 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii)No Ligation State
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, NO, ZN, PO4, GOL |
 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii)Co Ligation State, Q154A/Q155A/K156A Mutant
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, ZN, CMO, NA |

