Search Count: 17
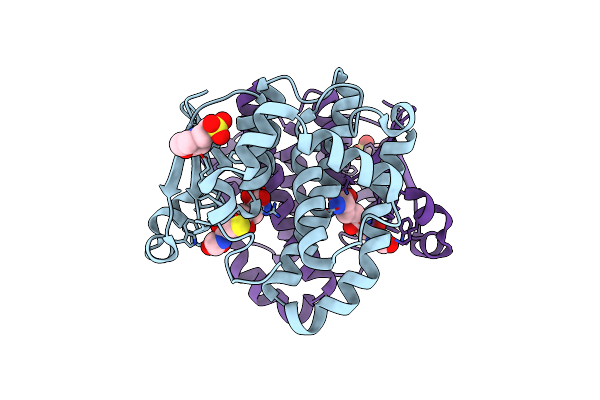 |
A Folding Mutant Of Human Class Pi Glutathione Transferase, Created By Mutating Aspartate 153 Of The Wild-Type Protein To Asparagine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-07-22 Classification: TRANSFERASE Ligands: MES, GSH |
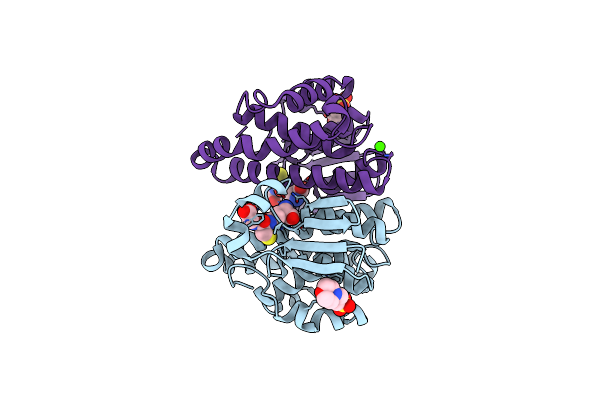 |
A Folding Mutant Of Human Class Pi Glutathione Transferase, Created By Mutating Aspartate 153 Of The Wild-Type Protein To Glutamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2003-07-22 Classification: TRANSFERASE Ligands: MES, GSH, CA |
 |
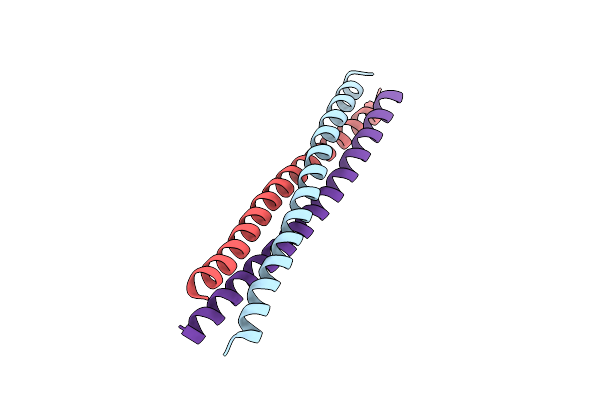
Crystal Structure Of A Novel Alanine-Zipper Trimer At 1.7 A Resolution, V13A,L16A,V20A,L23A,V27A,M30A,V34A Mutations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-06-17 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Crystal Structure Of A Novel Alanine-Zipper Trimer At 1.3 A Resolution, I6A,L9A,V13A,L16A,V20A,L23A,V27A,M30A,V34A,L48A,M51A Mutations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2003-06-17 Classification: MEMBRANE PROTEIN |
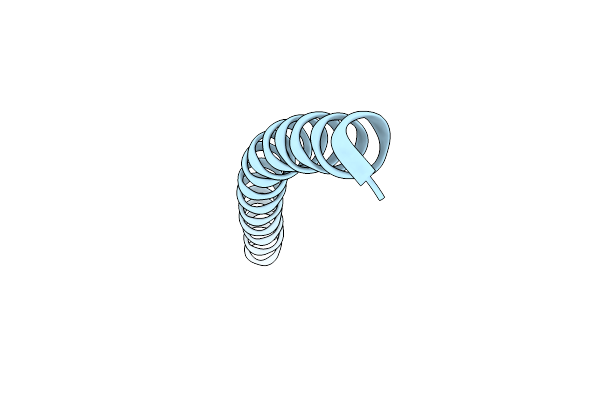 |
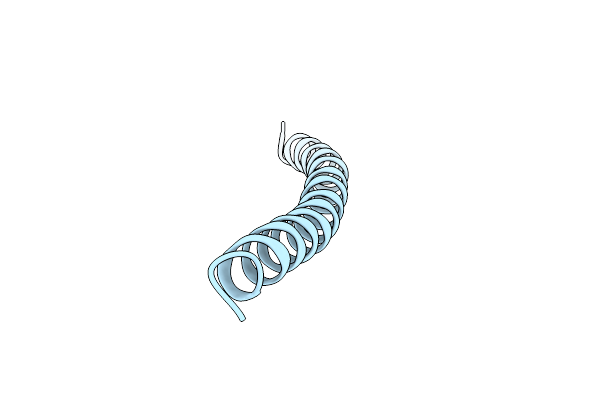
Core Side-Chain Packing And Backbone Conformation In Lpp-56 Coiled-Coil Mutants
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-06-28 Classification: MEMBRANE PROTEIN |
 |
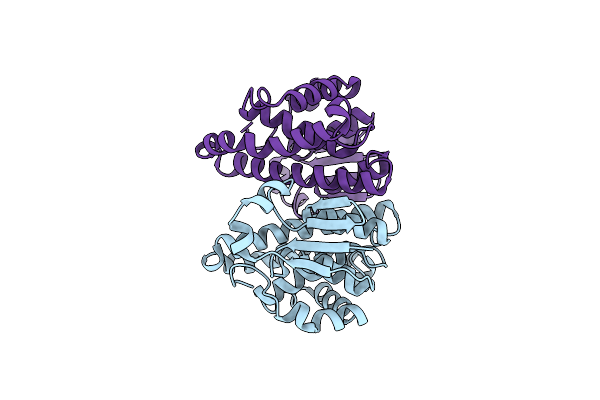
Core Side-Chain Packing And Backbone Conformation In Lpp-56 Coiled-Coil Mutants
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2002-06-28 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-10-18 Classification: TRANSFERASE |
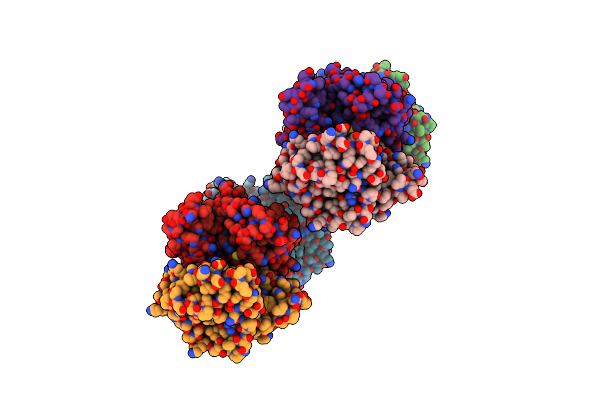 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2000-10-18 Classification: TRANSFERASE |
 |
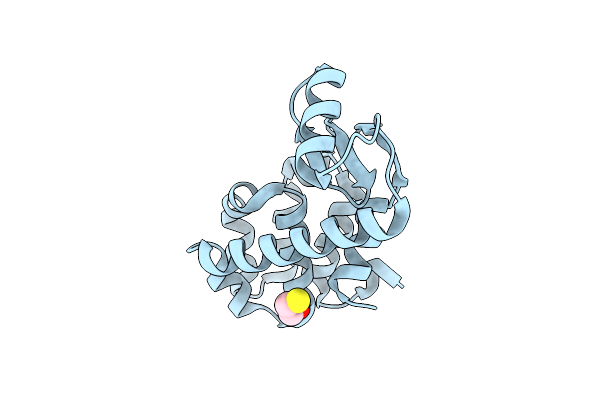
Core Structure Of The Outer Membrane Lipoprotein From Escherichia Coli At 1.9 Angstrom Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-06-28 Classification: MEMBRANE PROTEIN |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 1999-11-17 Classification: HYDROLASE Ligands: BME |
 |
 |
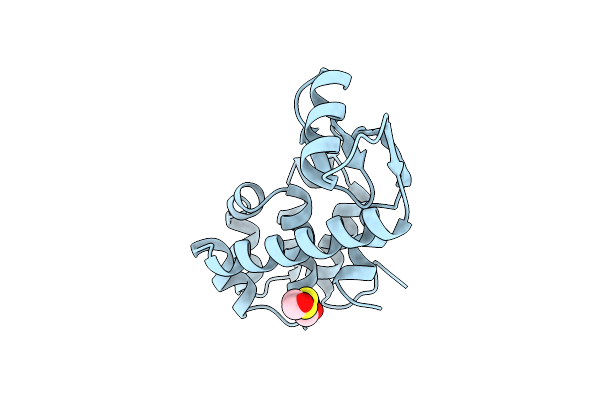
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
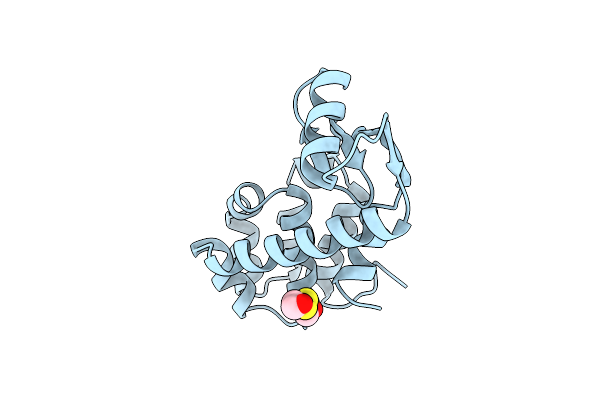
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Dissection Of Helix Capping In T4 Lysozyme By Structural And Thermodynamic Analysis Of Six Amino Acid Substitutions At Thr 59
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |

