Search Count: 94
 |
Organism: Pseudomonas pavonaceae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: LIGASE Ligands: G2P, MG, TA1 |
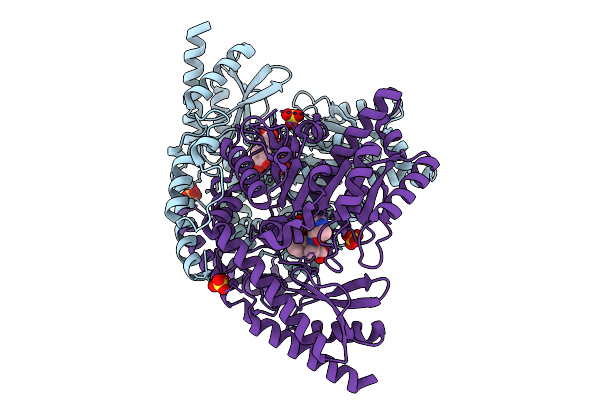 |
Re-Refined Of Crystal Structure Of Dopa Decarboxylase In Complex With The Inhibitor Carbidopa (1Js3) With Ketoenamine Form Of Carbidopa
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LYASE Ligands: A1BCS, SO4 |
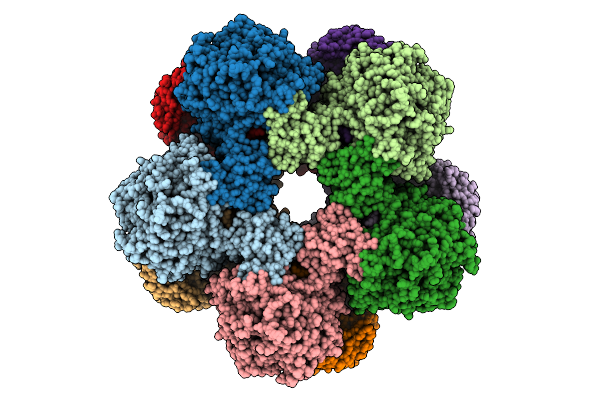 |
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei D-Hydrazino-Lysine Analog At 2.3 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LYASE Ligands: A1BD0 |
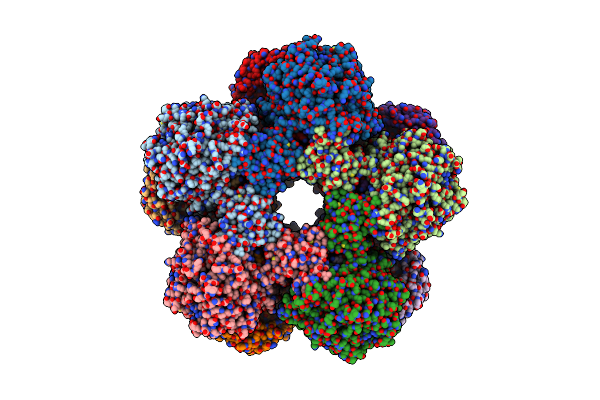 |
Cryoem Structure Of Holoenzyme Of Inducible Lysine Decarboxylase From Hafnia Alvei Holoenzyme At 2.19 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE |
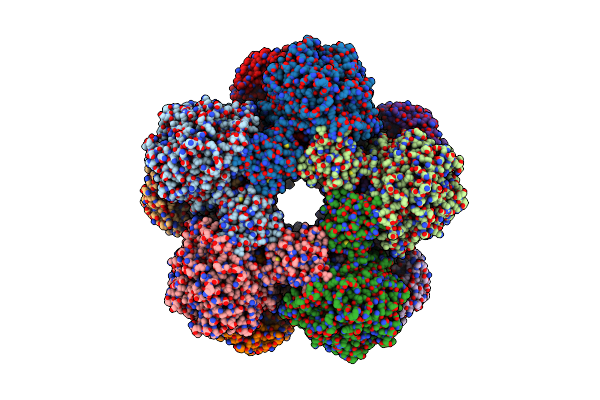 |
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei L-Hydrazino-Lysine Analog At 2.04 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE Ligands: A1BD1 |
 |
X-Ray Structure Of Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) Complex With Carbidopa At Physiological Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP, 142, PG4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Respiratory syncytial virus, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: VIRAL PROTEIN |
 |
Organism: Respiratory syncytial virus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: VIRAL PROTEIN |
 |
Stacks Of Nucleocapsid Rings Of The N1-370 Mutant Of The Human Respiratory Syncytial Virus
Organism: Human respiratory syncytial virus a strain long, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Toroid (Torc1 Organized In Inhibited Domains).
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: SIGNALING PROTEIN |
 |
Mosquitocidal Cry11Aa Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-Y449F Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-E583Q Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-F17Y Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Ba Determined At Ph 6.5 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar jegathesan
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Ba Determined At Ph 10.4 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar jegathesan
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-07-27 Classification: TOXIN Ligands: GOL |
 |
Organism: Providencia stuartii
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: LYASE |
 |
Organism: Providencia stuartii
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: LYASE |

