Search Count: 21
 |
Hsv-1 Origin Binding Protein In Complex With Double-Stranded Dna Recognition Sequence Oris With 6 Basepairs Removed From The At-Rich Region
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: DNA BINDING PROTEIN |
 |
Hsv-1 Origin Binding Protein In Complex With Double-Stranded Dna Recognition Sequence Oris And Non-Hydrolyzable Atp Analog
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: DNA BINDING PROTEIN Ligands: AGS, MG |
 |
Organism: Ranoidea citropa
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: ANTIMICROBIAL PROTEIN |
 |
Amyloid Fibril From The Antimicrobial Peptide Citropin 1-3 - Polymorph Nr.3
Organism: Ranoidea citropa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ANTIMICROBIAL PROTEIN |
 |
Amyloid Fibril From The Antimicrobial Peptide Citropin 1-3 - Polymorph Nr.2
Organism: Ranoidea citropa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ANTIMICROBIAL PROTEIN |
 |
Amyloid Fibril From The Antimicrobial Peptide Citropin 1-3 - Polymorph Nr.1L
Organism: Ranoidea citropa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ANTIMICROBIAL PROTEIN |
 |
Amyloid Fibril From The Antimicrobial Peptide Citropin 1-3 - Polymorph Nr.3L
Organism: Ranoidea citropa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ANTIMICROBIAL PROTEIN |
 |
Amyloid Fibril From The Antimicrobial Peptide Citropin 1-3 - Polymorph Nr.2L
Organism: Ranoidea citropa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ENDOCYTOSIS |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: ENDOCYTOSIS |
 |
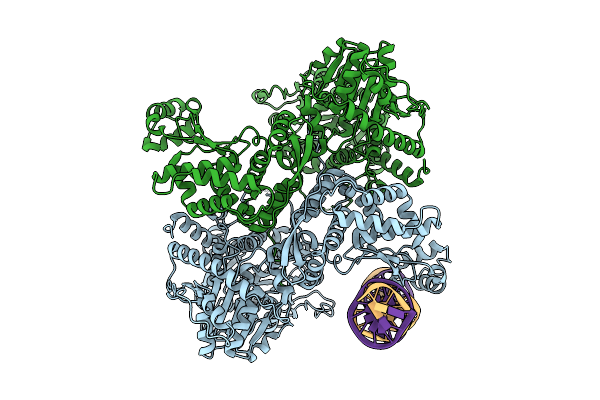
Hsv-1 Dna Polymerase-Processivity Factor Complex In Exonuclease State With 1-Bp Dna Mismatch
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.12 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: CA |
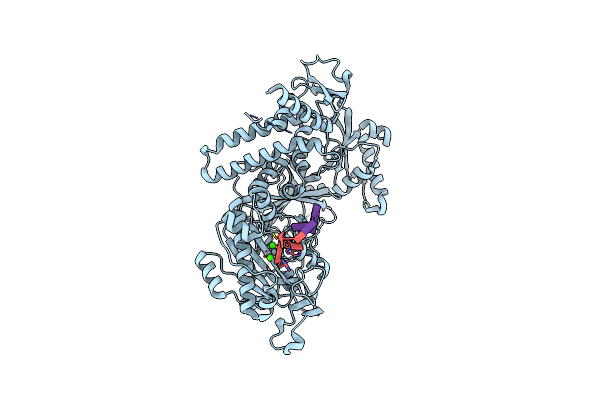 |
Hsv-1 Dna Polymerase-Processivity Factor Complex In Exonuclease State Active Site With 1-Bp Dna Mismatch
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.12 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: CA |
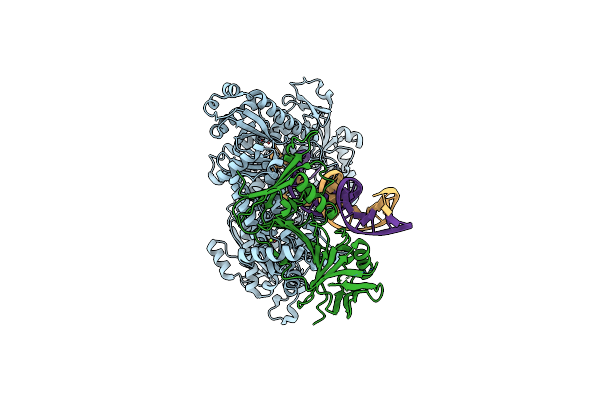 |
Hsv-1 Dna Polymerase-Processivity Factor Complex In Pre-Translocation State
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.41 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: MG |
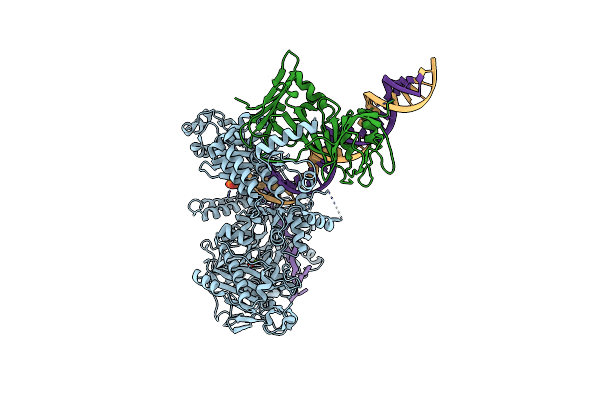 |
Hsv-1 Dna Polymerase-Processivity Factor Complex In Halted Elongation State
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.46 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: MG, DTP |
 |
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:1.87 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: CA |
 |
Hsv-1 Dna Polymerase-Processivity Factor Complex In Exonuclease State Active Site
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:1.90 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.08 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Human alphaherpesvirus 1 strain kos, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.46 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: CA |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2016-10-26 Classification: TRANSFERASE Ligands: ACT, MPD, LBV |
 |
Wild-Type Pas-Gaf Fragment From Deinococcus Radiodurans Bphp Collected At Lcls
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-10-26 Classification: TRANSFERASE Ligands: LBV, ACT |

