Search Count: 126
 |
Structure Of Meiothermus Ruber Mrub_1259 Lov Domain With N- And C-Terminal Alpha Helices (Mrlove)
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: DE NOVO PROTEIN Ligands: RET, SO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
True-Atomic Resolution Crystal Structure Of The Closed State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
True-Atomic Resolution Crystal Structure Of The Open State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, NA, 97N, OLC, LFA, GOL |
 |
Structure Of The Viral Channelrhodopsin Olpvr1 At Low Ph Obtained By Soaking
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, 97N, LFA, OLC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: GOL, PGE |
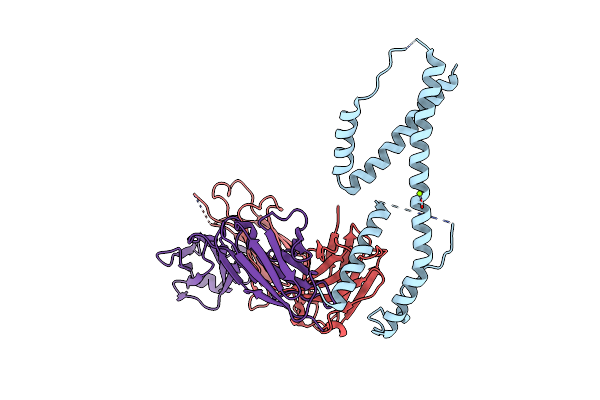 |
Crystal Structure Of The Fab Fragment Of The Anti-Il-6 Antibody I9H In Complex With A Domain-Swapped Il-6 Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Crystal Structure Of The Fab Fragment Of The Anti-Il-6 Antibody 68F2 In Complex With A Domain-Swapped Il-6 Dimer
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: SO4 |
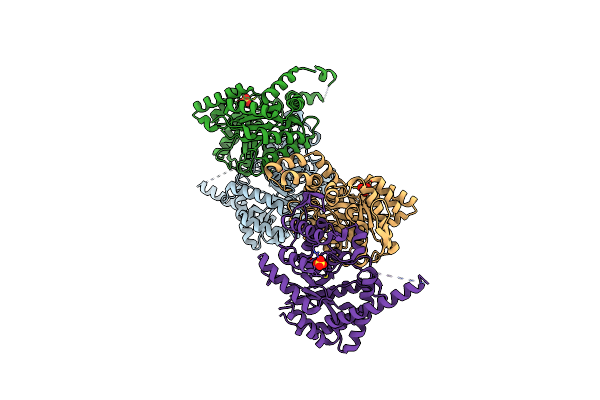 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-02-07 Classification: LUMINESCENT PROTEIN Ligands: SO4 |
 |
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-08-16 Classification: FLUORESCENT PROTEIN Ligands: CYC |

