Search Count: 6
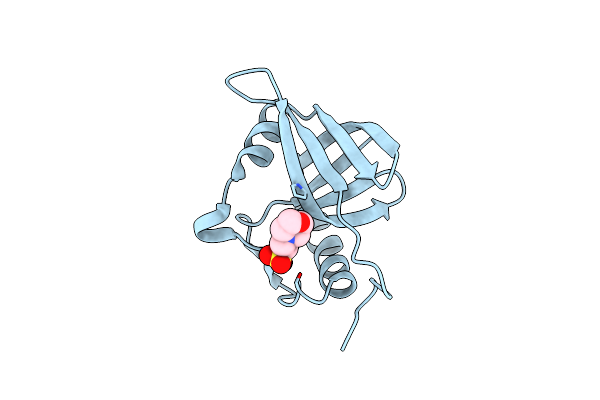 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
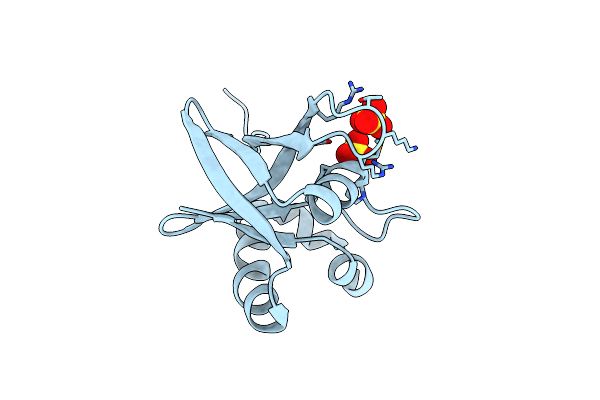
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Nitroreductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Reductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42, SO4 |

