Search Count: 19
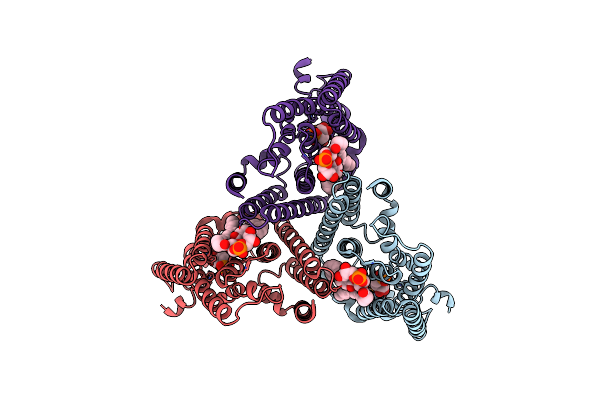 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: PRP, MG, PGW |
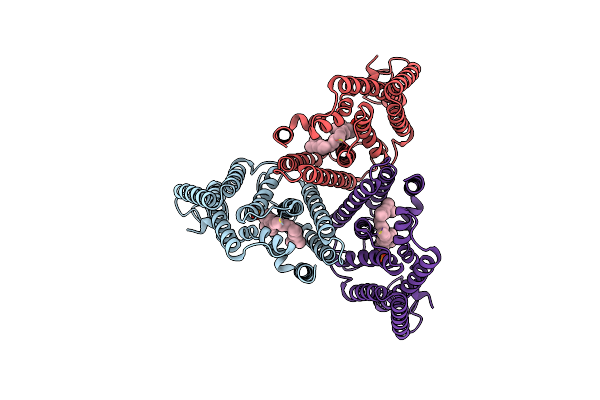 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: DSL |
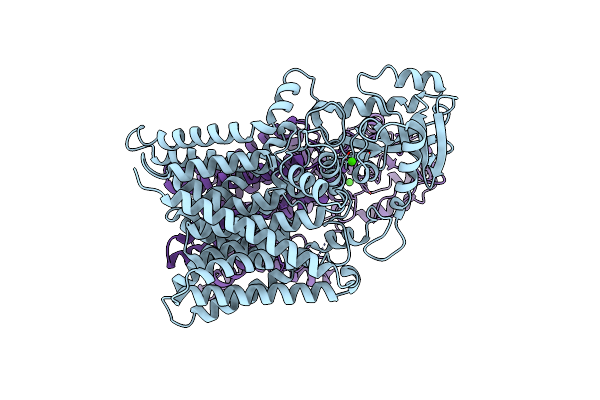 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: CA |
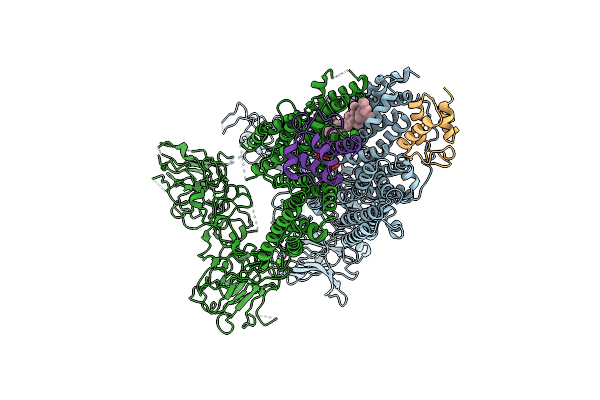 |
Mycobacterium Smegmatis Arabinosyltransferase Complex Embb2-Acpm2 In Substrate Dpa Bound Asymmetric "Active State"
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-05-27 Classification: TRANSFERASE Ligands: CA, F8L |
 |
Mycobacterium Smegmatis Arabinosyltransferase Complex Embb2-Acpm2 In Symmetric "Resting State"
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-05-27 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Mycobacterium Smegmatis Arabinosyltransferase Emba-Embb-Acpm2 In Complex With Ethambutol
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-04-29 Classification: TRANSFERASE Ligands: CDL, F8L, CA, PNS, PO4, 95E |
 |
Cryo-Em Structure Of Mycobacterium Smegmatis Arabinosyltransferase Embc2-Acpm2 In Complex With Ethambutol
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155), Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-04-29 Classification: TRANSFERASE Ligands: CA, PO4, 95E, PN7 |
 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Arabinosyltransferase Emba-Embb-Acpm2 In Complex With Ethambutol
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-04-29 Classification: TRANSFERASE Ligands: CA, 95E, DSL, CDL |
 |
Cryo-Em Structure Of Mycobacterium Smegmatis Arabinosyltransferase Emba-Embb-Acpm2 In Complex With Di-Arabinose.
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2020-04-29 Classification: TRANSFERASE Ligands: F8L, CDL, CA, PO4, PNS |
 |
Crystal Structure Of Arabinosyltransferase Embc2-Acpm2 Complex From Mycobacterium Smegmatis Complexed With Di-Arabinose
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2020-04-29 Classification: TRANSFERASE Ligands: CA, PO4, BXY |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2016-09-21 Classification: TRANSFERASE |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN Ligands: 5F9 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN Ligands: 5FF |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2016-01-20 Classification: LYASE Ligands: G59 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN Ligands: 5FK |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN Ligands: G51 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-01-20 Classification: LIPID BINDING PROTEIN Ligands: G7A |
 |
Biochemical And Structural Characterization Of Mycobacterial Aspartyl-Trna Synthetase Asps, A Promising Tb Drug Target
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-11-19 Classification: LIGASE Ligands: 3SY, FMT |

