Search Count: 169
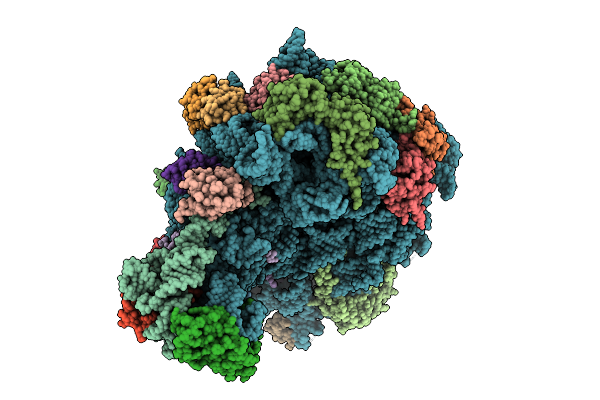 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
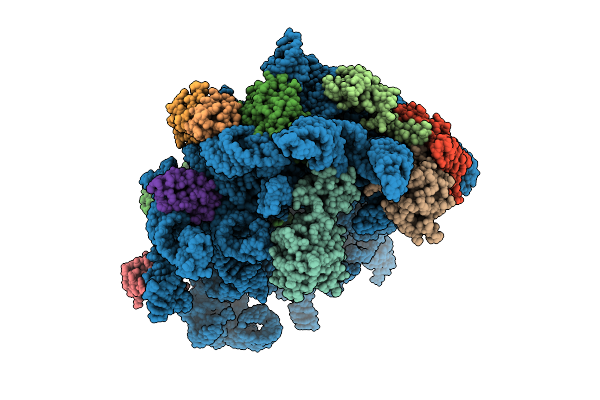 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
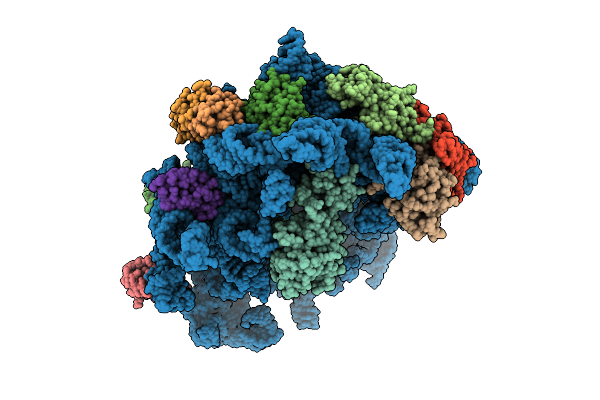 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
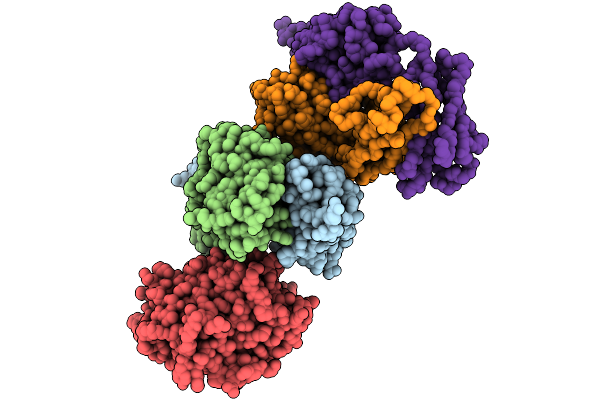 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-08-27 Classification: PROTEIN BINDING |
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-06-25 Classification: PEPTIDE BINDING PROTEIN Ligands: TEW, SO4 |
 |
1.8 A Resolution Structure Of The Photosystem I Assembly Intermediate Lacking Stromal Subunits.
Organism: Synechocystis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: PHOTOSYNTHESIS Ligands: LHG, CL0, CLA, PQN, BCR, ECH, LMG, LMT, CL, SF4, 5X6 |
 |
Structure And Interactions Of Hiv-1 Gp41 Chr-Nhr Reverse Hairpin Constructs Reveal Molecular Determinants Of Antiviral Activity
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: GOL, SO4 |
 |
Structure And Interactions Of Hiv-1 Gp41 Chr-Nhr Reverse Hairpin Constructs Reveal Molecular Determinants Of Antiviral Activity
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: SO4, GOL |
 |
Structure And Interactions Of Hiv-1 Gp41 Chr-Nhr Reverse Hairpin Constructs Reveal Molecular Determinants Of Antiviral Activity
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Structure And Interactions Of Hiv-1 Gp41 Chr-Nhr Reverse Hairpin Constructs Reveal Molecular Determinants Of Antiviral Activity
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Structure And Interactions Of Hiv-1 Gp41 Chr-Nhr Reverse Hairpin Constructs Reveal Molecular Determinants Of Antiviral Activity
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN |
 |
Organism: Brucella abortus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-01-22 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Halofuginone
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: HFG, ZN, CL, GOL |
 |
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Halofuginone And Atp Analogue
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: ANP, HFG, MG, ZN |
 |
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Two Inhibitors (Halofuginone And L95)
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: ZN, HFG, JE6, BR, GOL, CL |
 |
Crystal Structure Of Hla-A*02:01/Ny-Eso-1 (Sllmwitqv) And A Target Specific Tracer-I
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-20 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of Apo- Exo-Beta-(1,3)-Glucanase From Aspergillus Oryzae (Aobgl)
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1L0T, NA |
 |
Crystal Structure Of Exo-Beta-(1,3)-Glucanase From Aspergillus Oryzae (Aobgl) As A Complex With Cellobiose
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: BGC, NA |
 |
High-Resolution Crystal Structure Of Exo-Beta-(1,3)-Glucanase From Aspergillus Oryzae (Aobgl) As A Complex With Glucose
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: EDO, GOL, BGC, CL, NA |
 |
Organism: Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: MEMBRANE PROTEIN |

