Search Count: 126
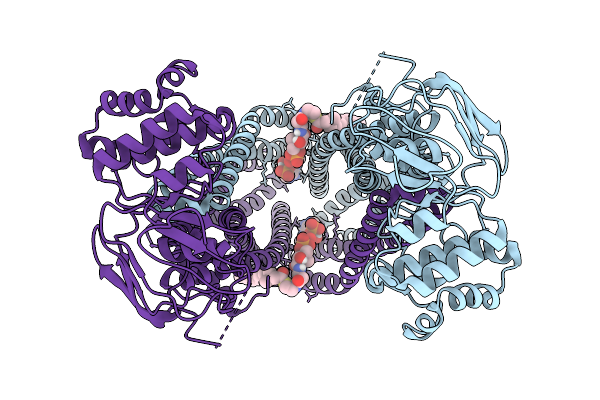 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: A1CAN |
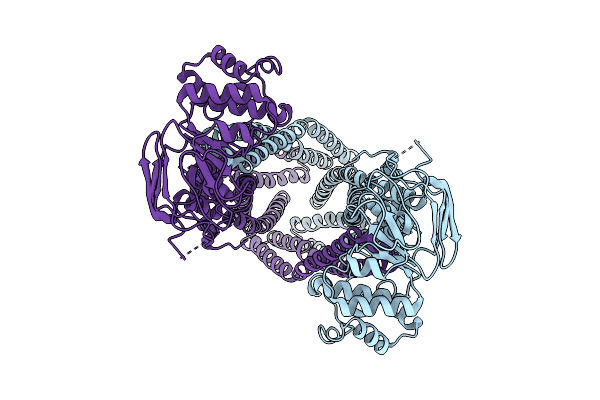 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Finding The Exit Route Of Hydrogen Peroxide From The Manganese Superoxide Dismutase (Mnsod) Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: K, PO4, MN |
 |
Finding The Exit Route Of Hydrogen Peroxide From The Manganese Superoxide Dismutase (Mnsod) Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: PEO, PO4, K, MN |
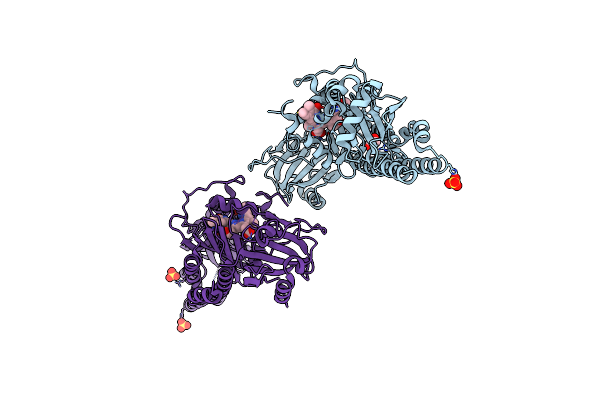 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
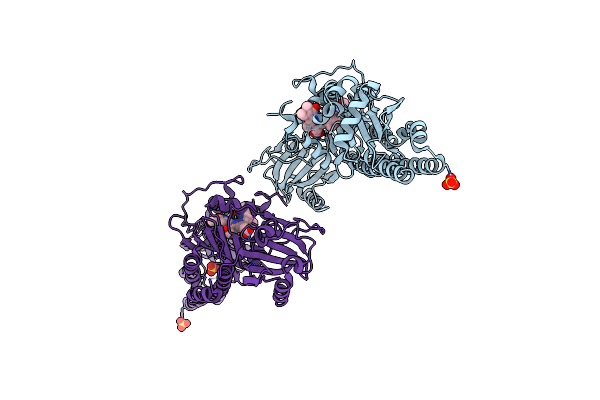 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Organism: Serendipita indica
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Serendipita indica
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN |
 |
Organism: Serendipita indica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: PO4, BNG |
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN, PEO |
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.50 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Organism: Homo sapiens
Method: NEUTRON DIFFRACTION Resolution:2.28 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN |
 |
X-Ray Counterpart To The Neutron Structure Of Peroxide-Soaked Tyr34Phe Mnsod
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: MN, PEO, PO4, K |

